# A tibble: 344 × 8
species island bill_length_mm bill_depth_mm flipper_length_mm body_mass_g
<fct> <fct> <dbl> <dbl> <int> <int>
1 Adelie Torgersen 39.1 18.7 181 3750
2 Adelie Torgersen 39.5 17.4 186 3800
3 Adelie Torgersen 40.3 18 195 3250
4 Adelie Torgersen NA NA NA NA
5 Adelie Torgersen 36.7 19.3 193 3450
6 Adelie Torgersen 39.3 20.6 190 3650
7 Adelie Torgersen 38.9 17.8 181 3625
8 Adelie Torgersen 39.2 19.6 195 4675
9 Adelie Torgersen 34.1 18.1 193 3475
10 Adelie Torgersen 42 20.2 190 4250
# ℹ 334 more rows
# ℹ 2 more variables: sex <fct>, year <int>10: data visualization
STA35B: Statistical Data Science 2
Purpose of visualizing data
- Exploratory data analysis
- Presenting findings to others
Affects how much effort to put in
- If just exploring data, a plot doesn’t need to look pretty if you can interpret it
- If high-stakes presentation (e.g. for job interview, raise, promotion, etc), might need to add many bespoke features (not focus of this class)
Exploratory data analysis
Cycle through the following:
Generate questions about your data.
Search for answers by visualizing, transforming, and modelling your data.
Use what you learn to refine your questions and/or generate new questions
Requires creativity and critical thinking. Two question categories:
- What type of variation occurs within each variable?
- Mean, standard deviation, skewness, etc
- What type of covariation occurs between variables?
- How does height vary with weight, etc
ggplot2
Visualization
We’ll see how to create beautiful visualizations using ggplot2.
Basic structure of ggplot2
ggplot() constructs the initial plot.
- The first argument of
ggplot()is the data set for the plot.- The data set must be a data frame.
ggplot(data = mpg)creates an empty plot.
You then add one or more layers to ggplot() using +.
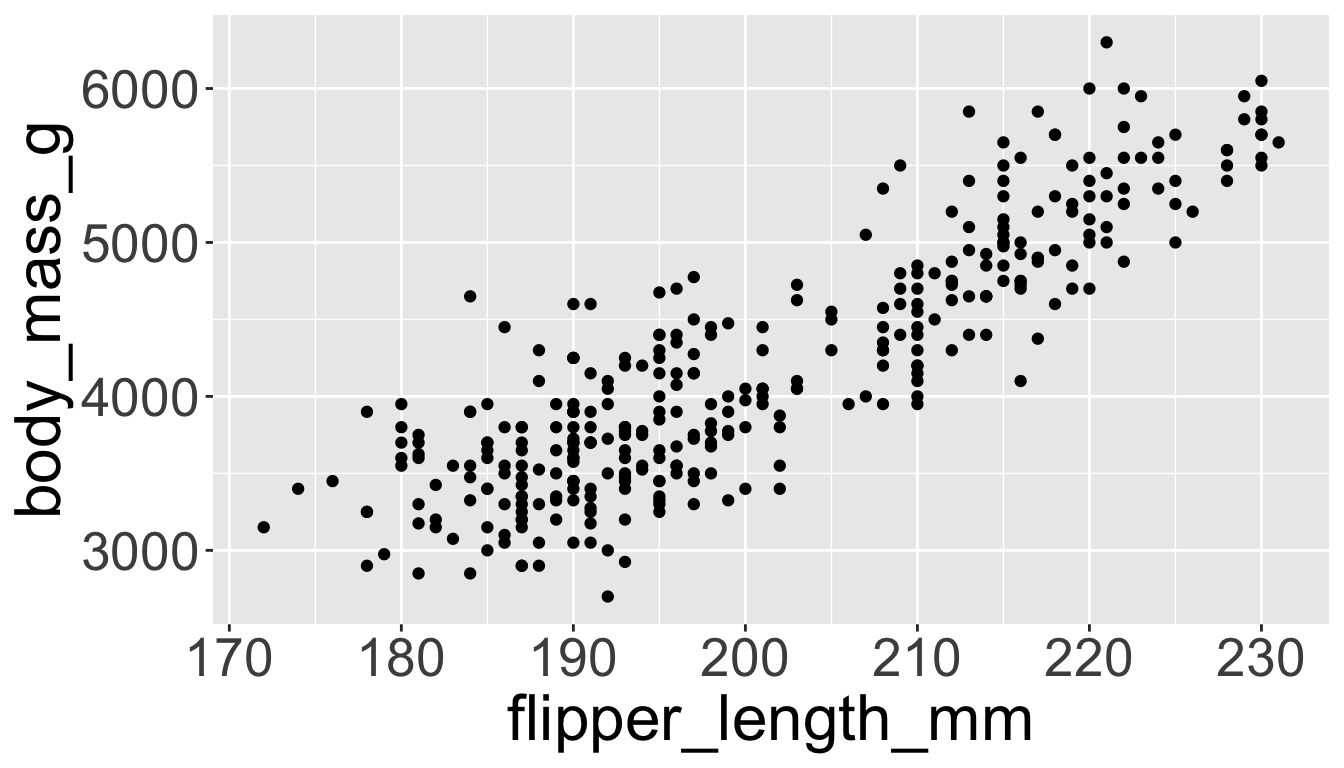
geomfunctions add a geometrical object to the plot.geom_point(),geom_smooth(),geom_histogram(),geom_boxplot(), etc.
Creating a ggplot
Creating a ggplot
Creating a ggplot
- Start with function
ggplot() - Add global aesthetics (i.e., aesthetics applied to every layer in plot).
- Add layers.
- Display data using geom: geometrical object used to represent data
geom_bar(): bar chart;geom_line(): lines;geom_boxplot(): boxplot;geom_point(): scatterplot
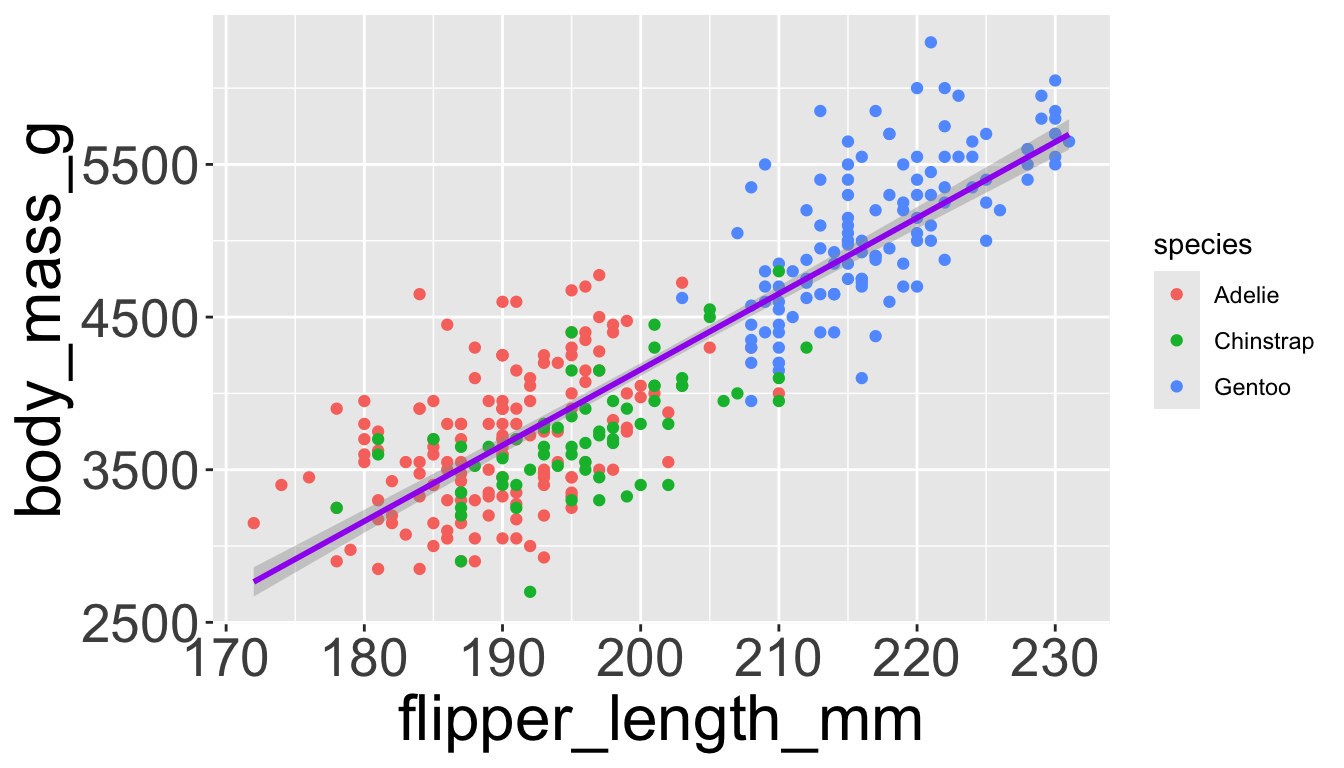
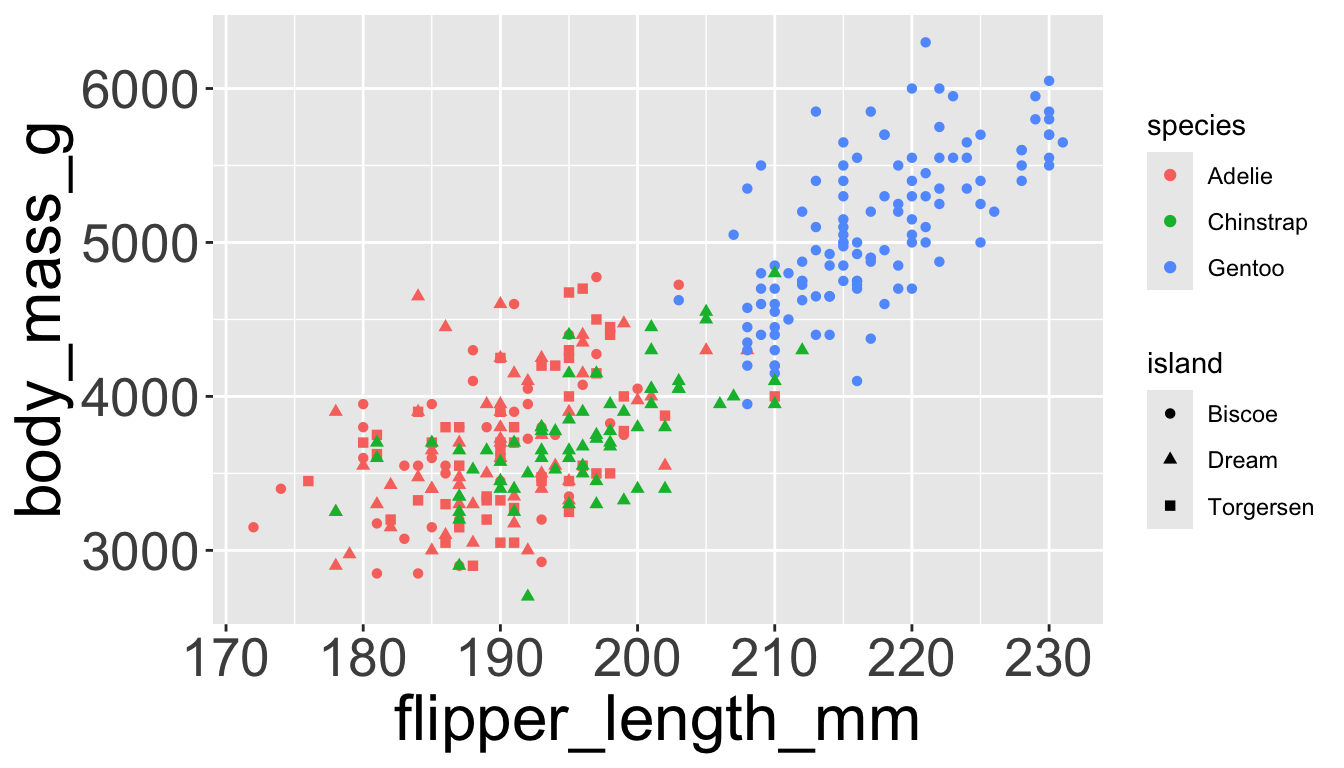
Adding aesthetics and layers
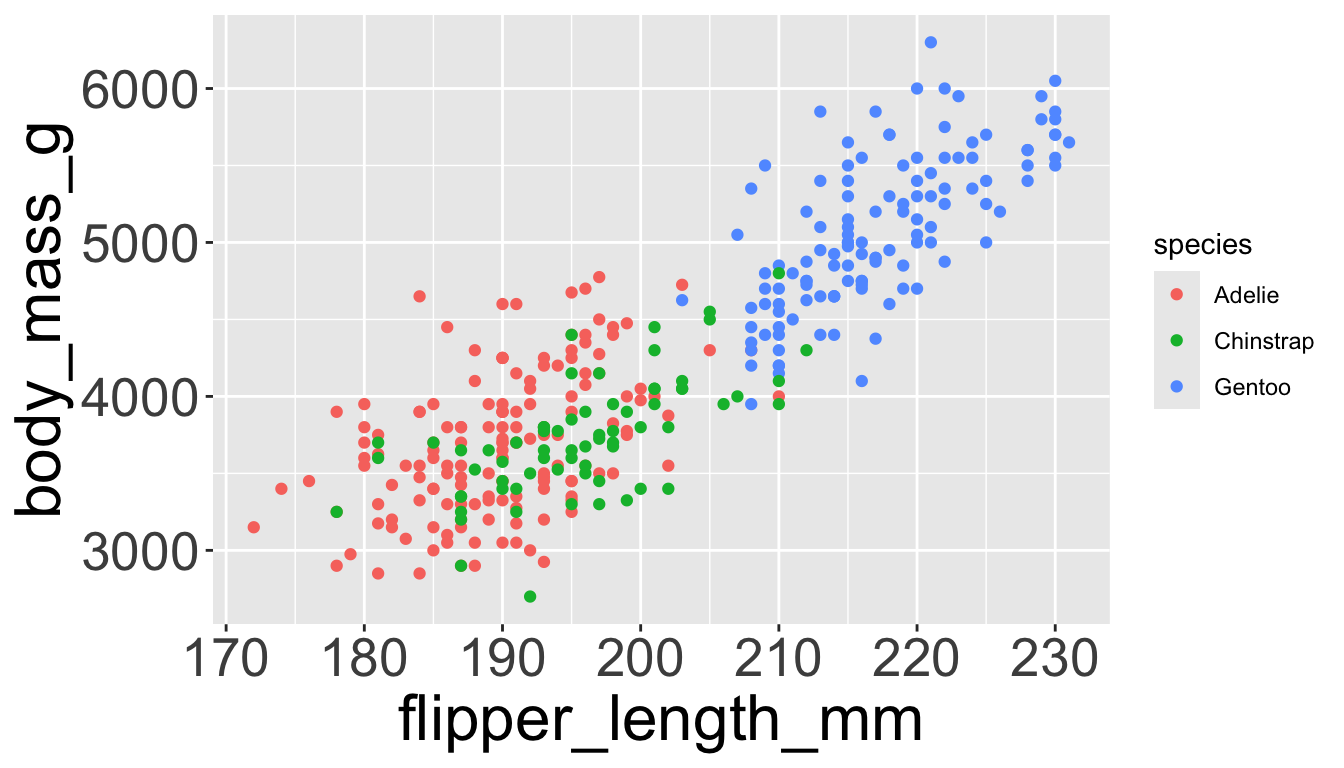
We can have aesthetics change as a function of variables inside the tibble
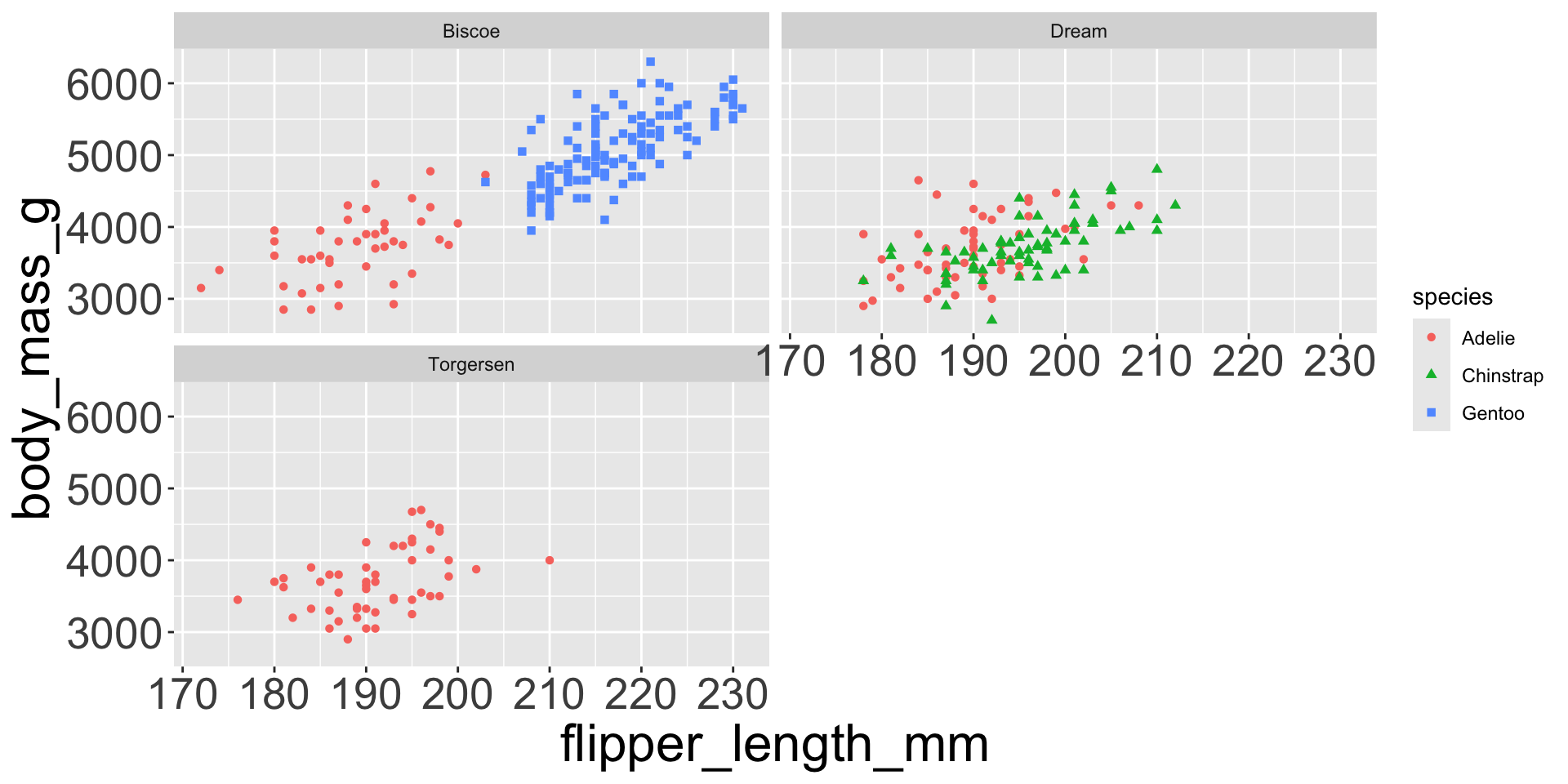
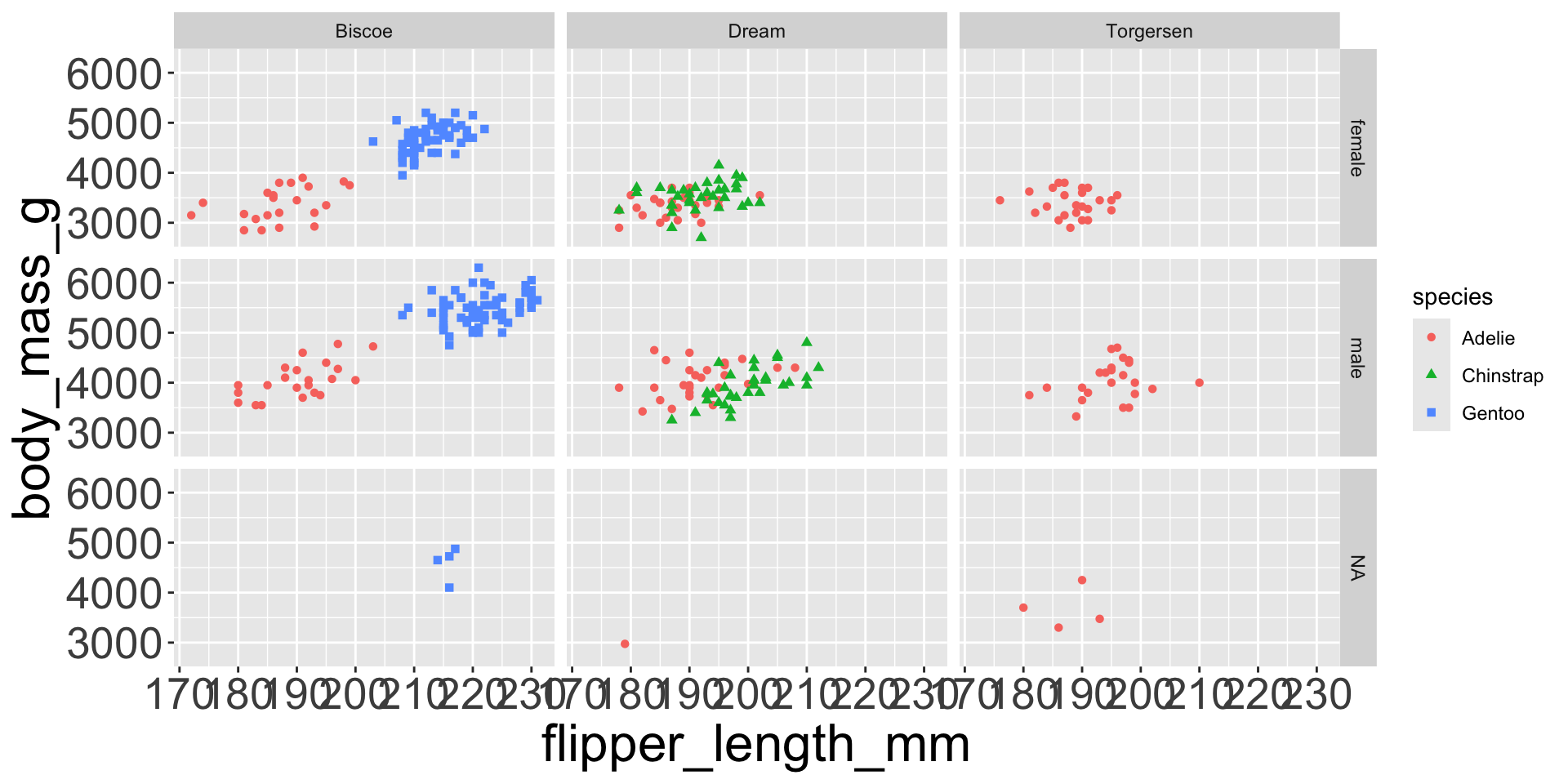
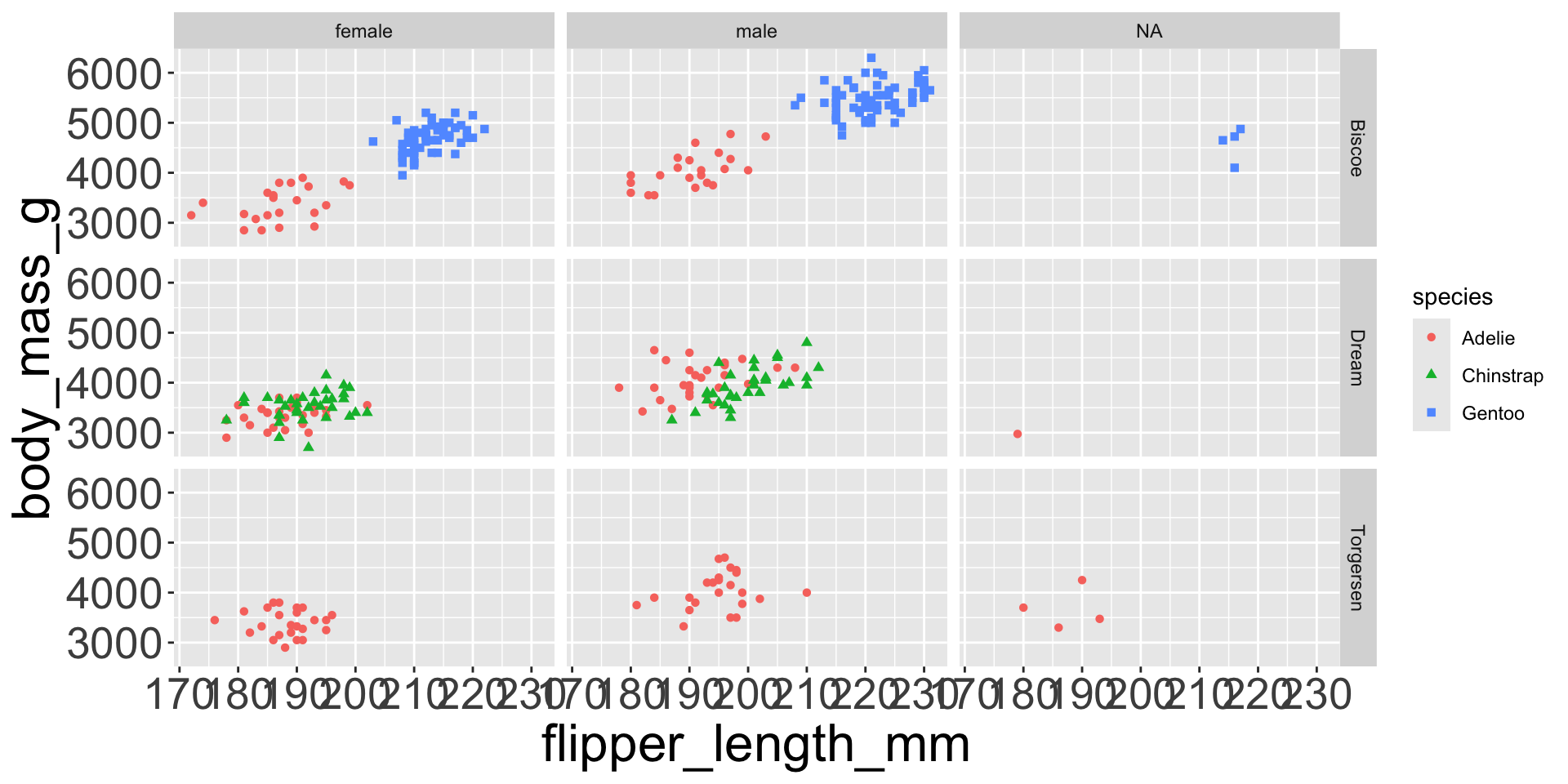
- e.g. we can differentiate penguin species via colors
- When a categorical variable is mapped to an aesthetic, each unique level of the variable (here: species) gets assigned a unique aesthetic value (here: unique color)
Adding aesthetics and layers
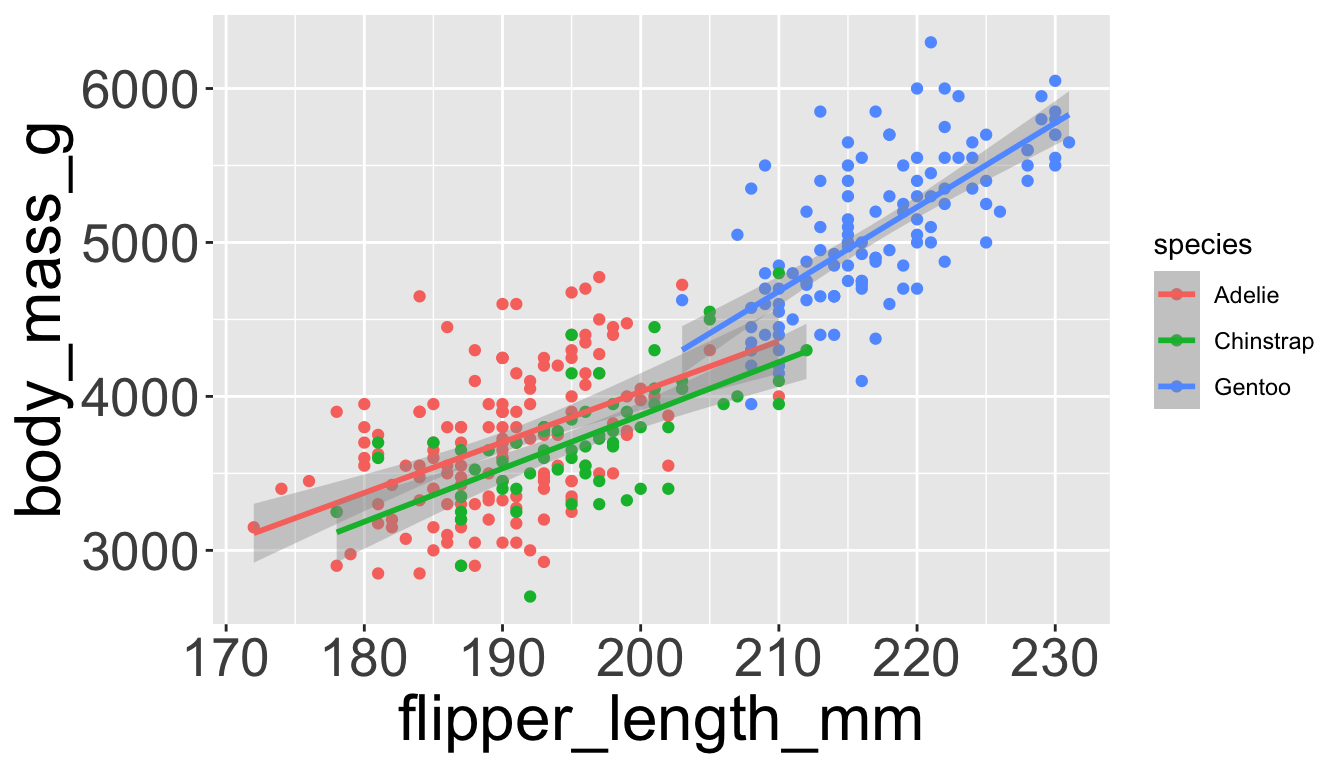
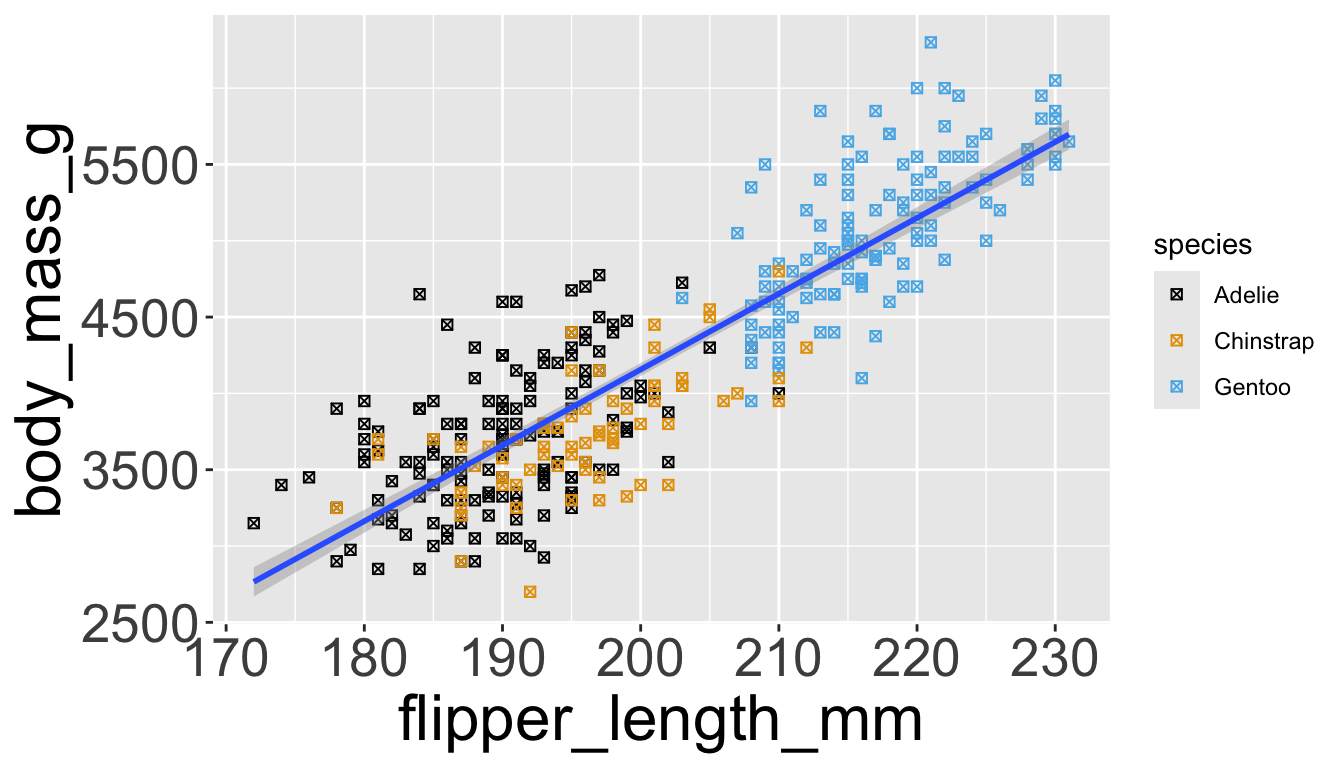
Let’s add a new layer, geom_smooth(method="lm"), which visualizes line of best fit based on a linear model
- When an aesthetic mapping is added inside
ggplot(), it is applied to all layers.- So
color=speciesinsideggplot()will group all penguins by species. - We now have a line for each species (not one global line).
- So
Adding aesthetics and layers
Let’s add a new layer, geom_smooth(method="lm"), which visualizes line of best fit based on a linear model
- When an aesthetic mapping is added inside a layer, it is applied to just that layer.
- So
color=speciesinsidegeom_point()will group all penguins by species only for that layer. - We now have one global line for all penguins.
- So
Adding aesthetics and layers
Adding aesthetics and layers
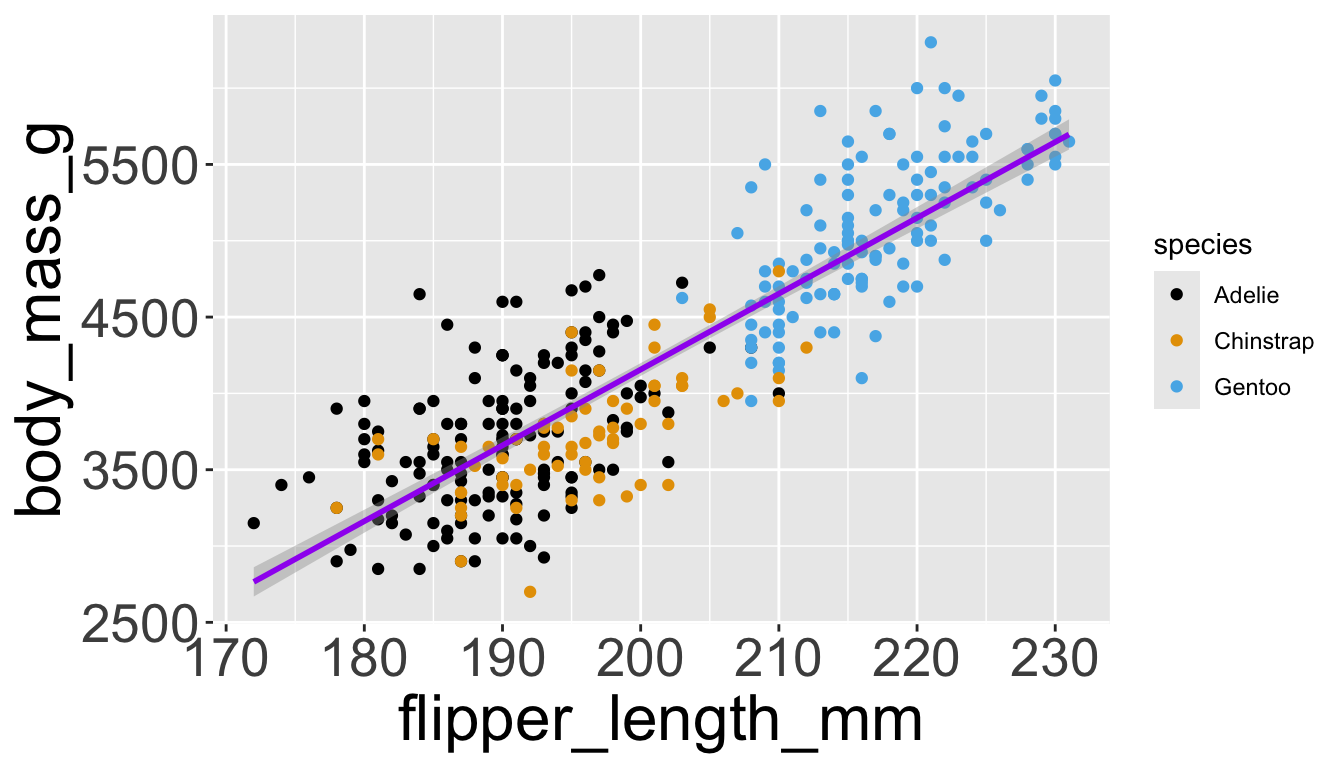
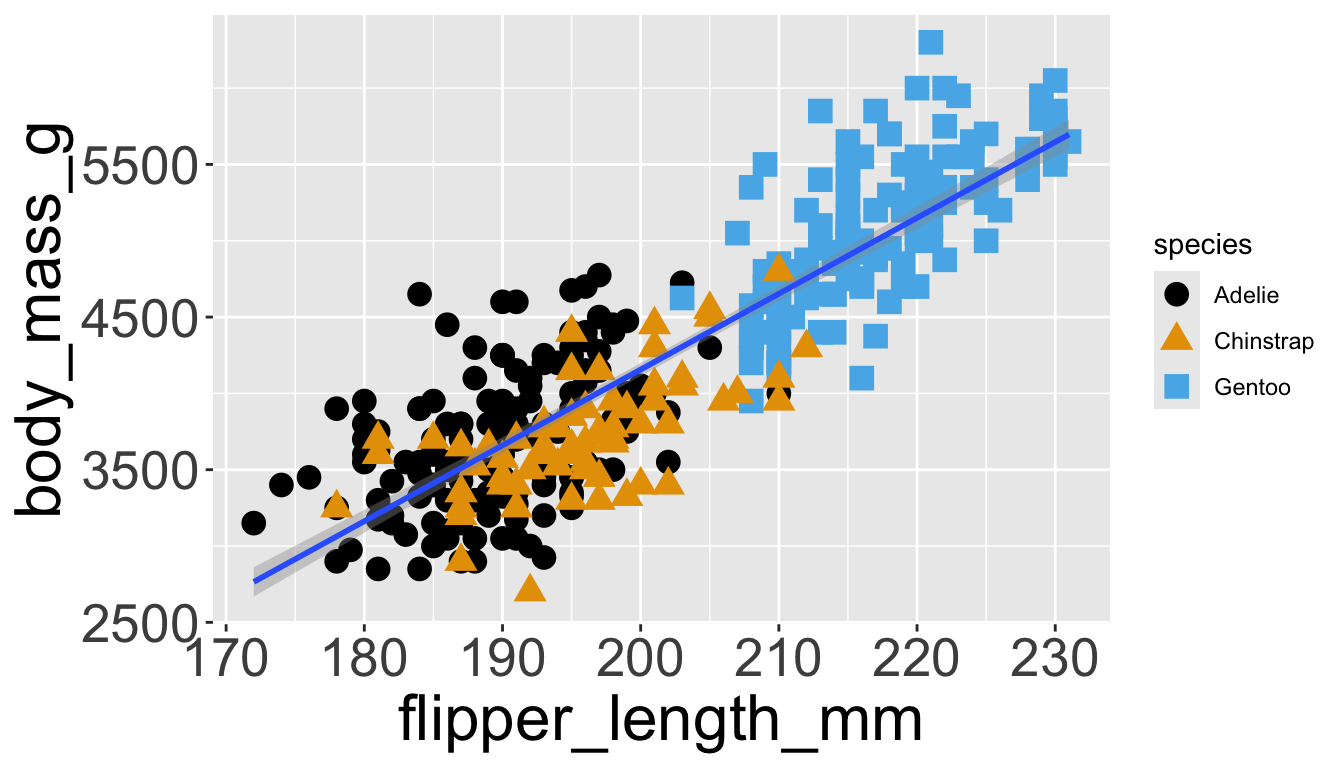
Let’s further differentiate different species via shapes.
- We can specify this in a local aesthetic mapping of points using
shape= - The legend will be updated to show this too!
Adding aesthetics and layers
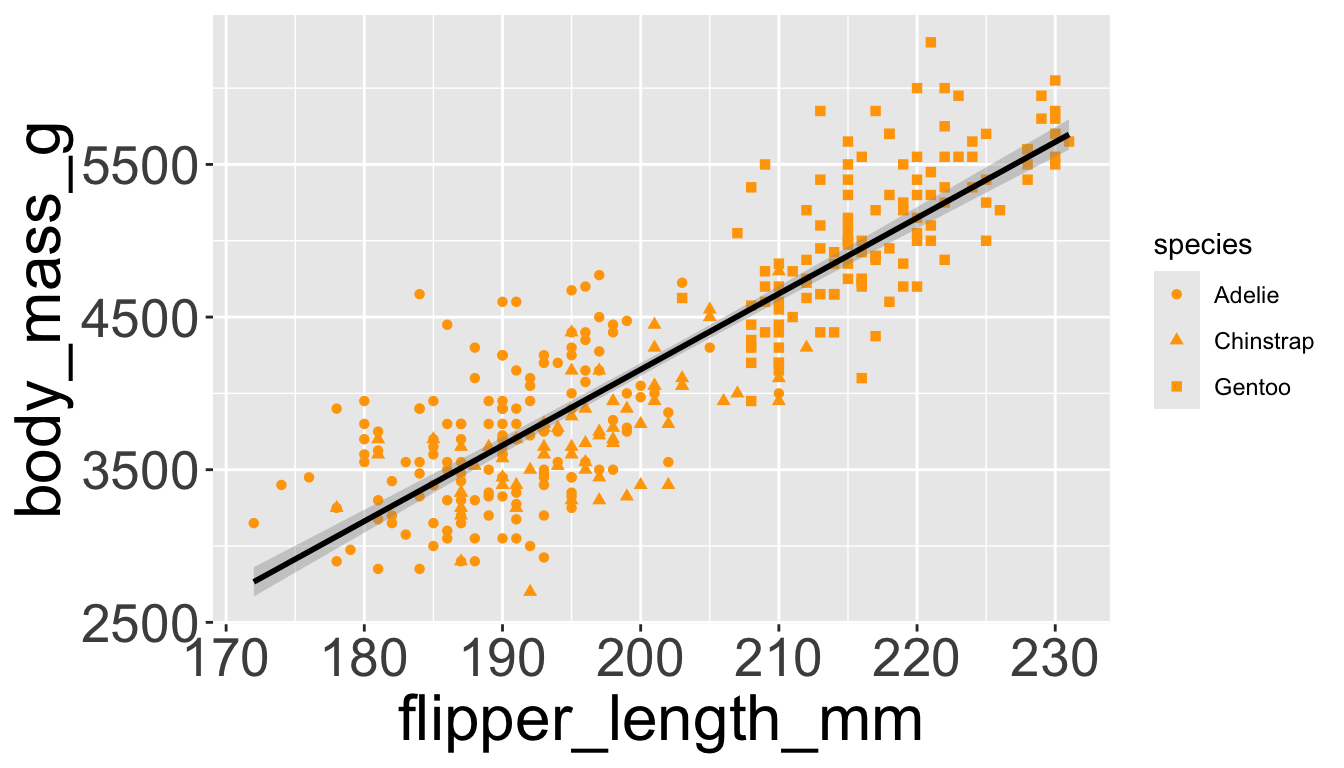
Let’s further differentiate different species via shapes.
- We can make all points the same color by specifying
color=outside ofaes()
Adding aesthetics and layers
Adding aesthetics and layers
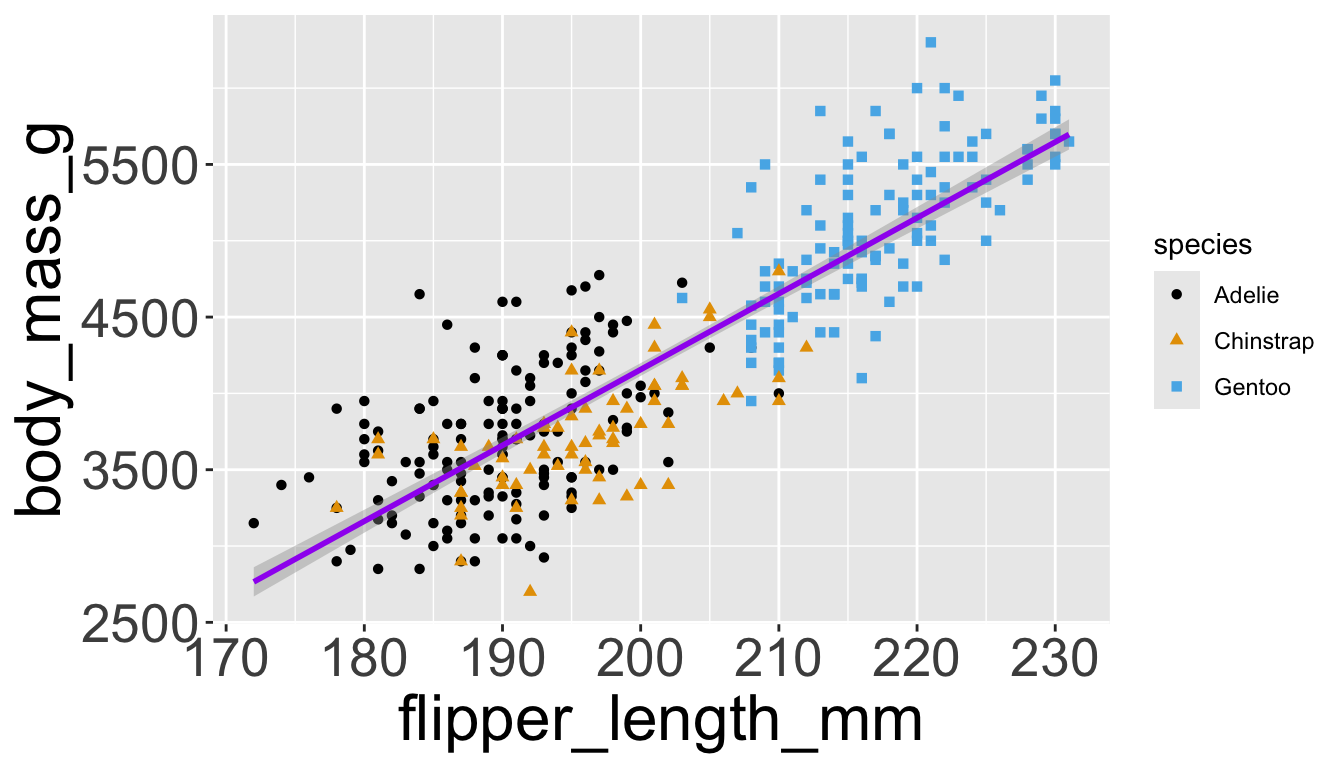
Let’s further differentiate different species via shapes.
- We can also specify
shape=outside ofaes()
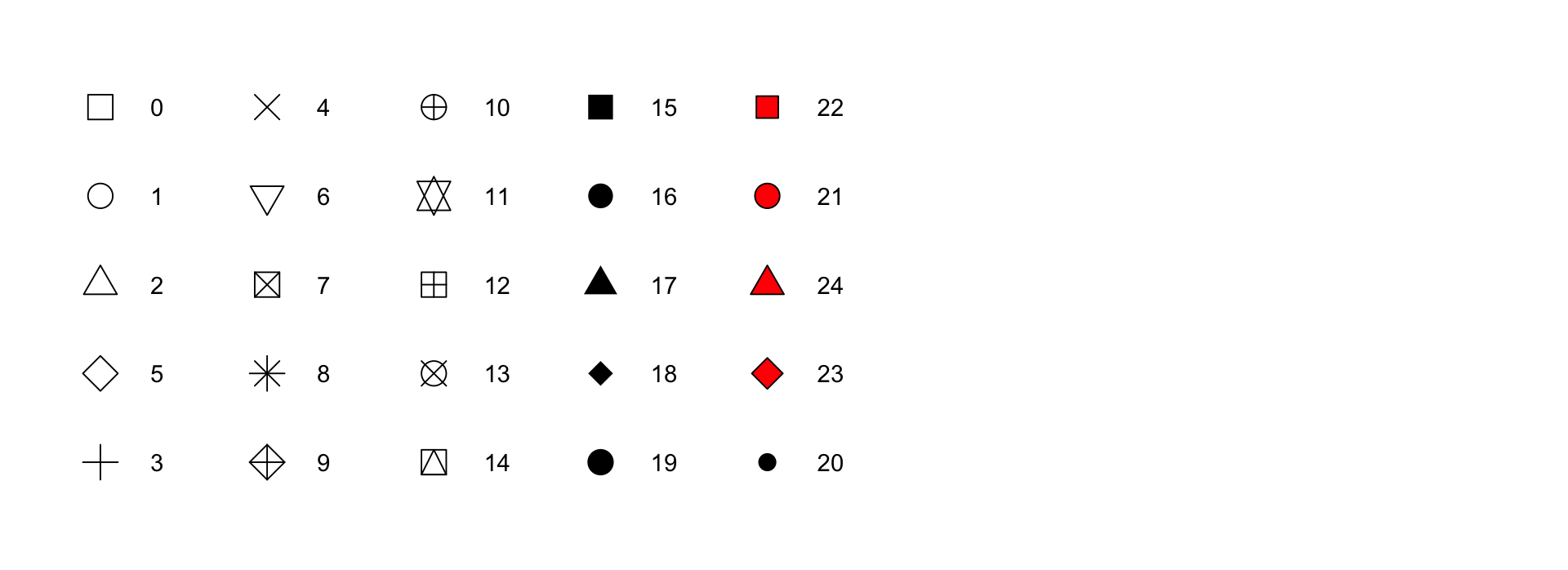
color and fill aesthetics. The hollow shapes (0–14) have a border determined by color; the solid shapes (15–20) are filled with color; the filled shapes (21–24) have a border of color and are filled with fill.
Adding aesthetics and layers
Now just need to add title and axis labels
penguins |>
ggplot(aes(x = flipper_length_mm,
y = body_mass_g)) +
geom_point(aes(color = species,
shape = species)) +
geom_smooth(method = "lm") +
labs(
title = "Body mass and flipper length",
subtitle = "Dimensions for Adelie, Chinstrap, and Gentoo Penguins",
x = "Flipper length (mm)", y = "Body mass (g)",
color = "Species", shape = "Species"
) +
scale_color_colorblind()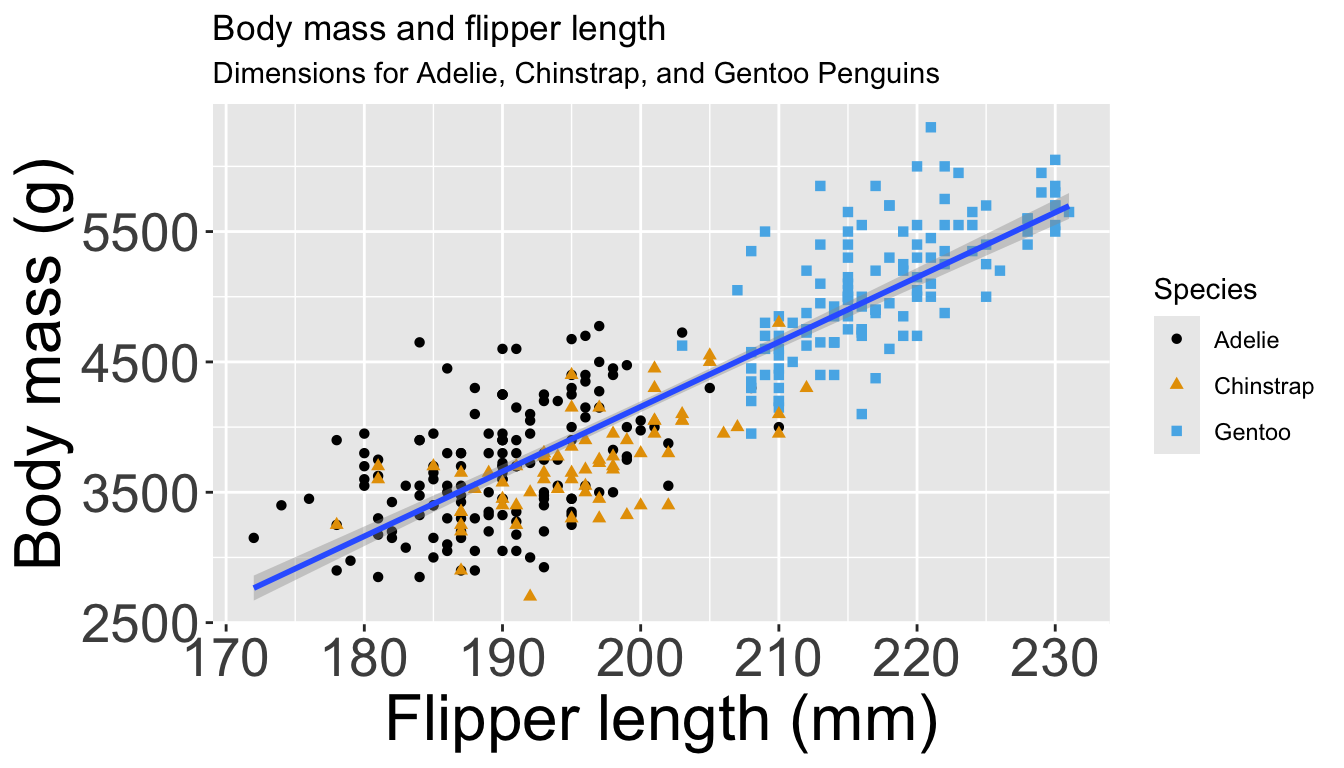
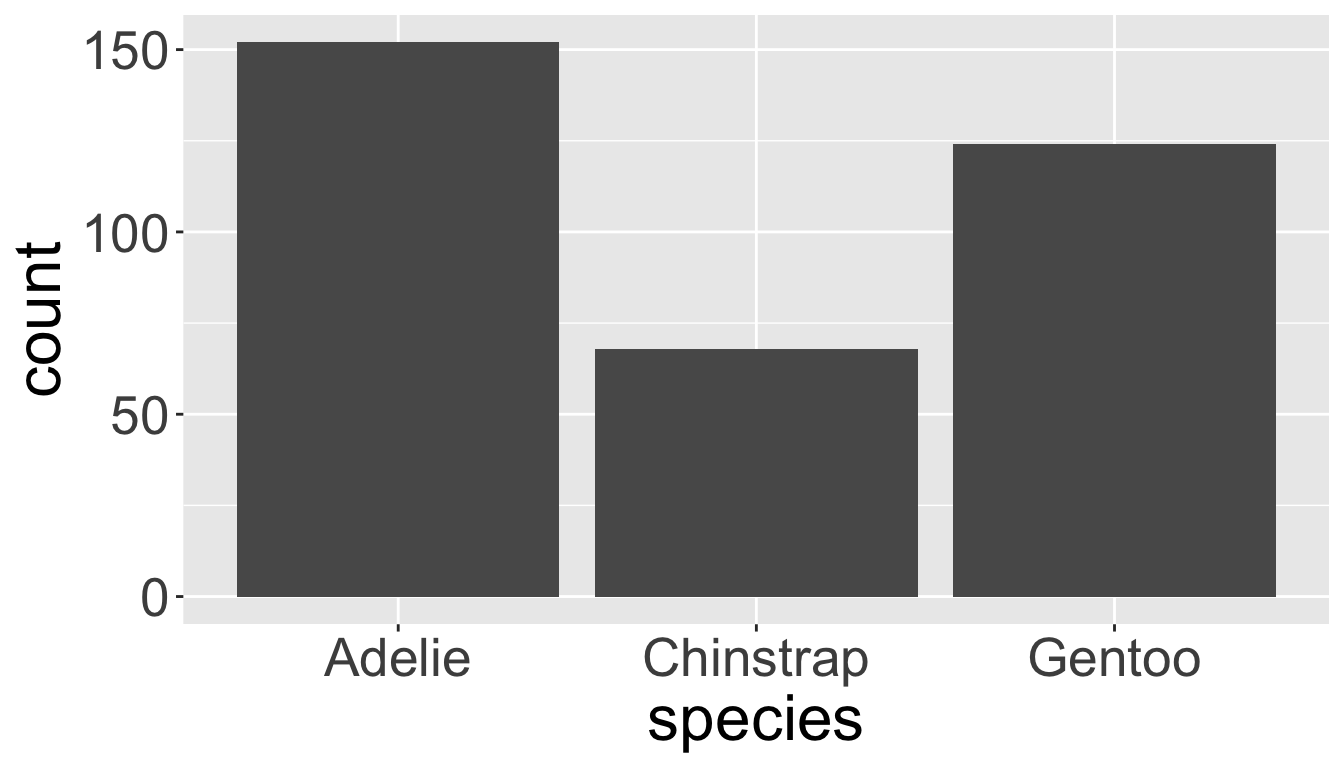
Visualizing distributions
Categorical variables take only one of a finite set of values
- Bar charts are useful for visualizing categorical variables
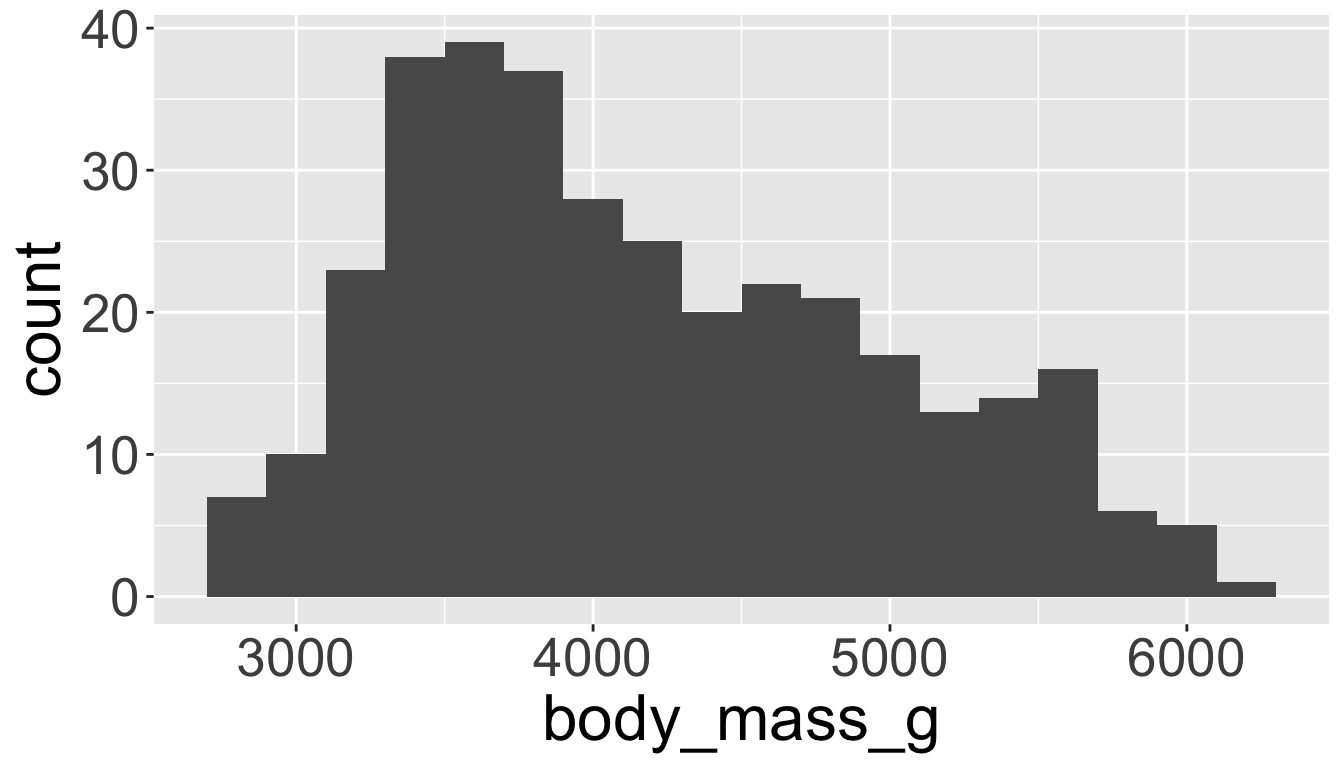
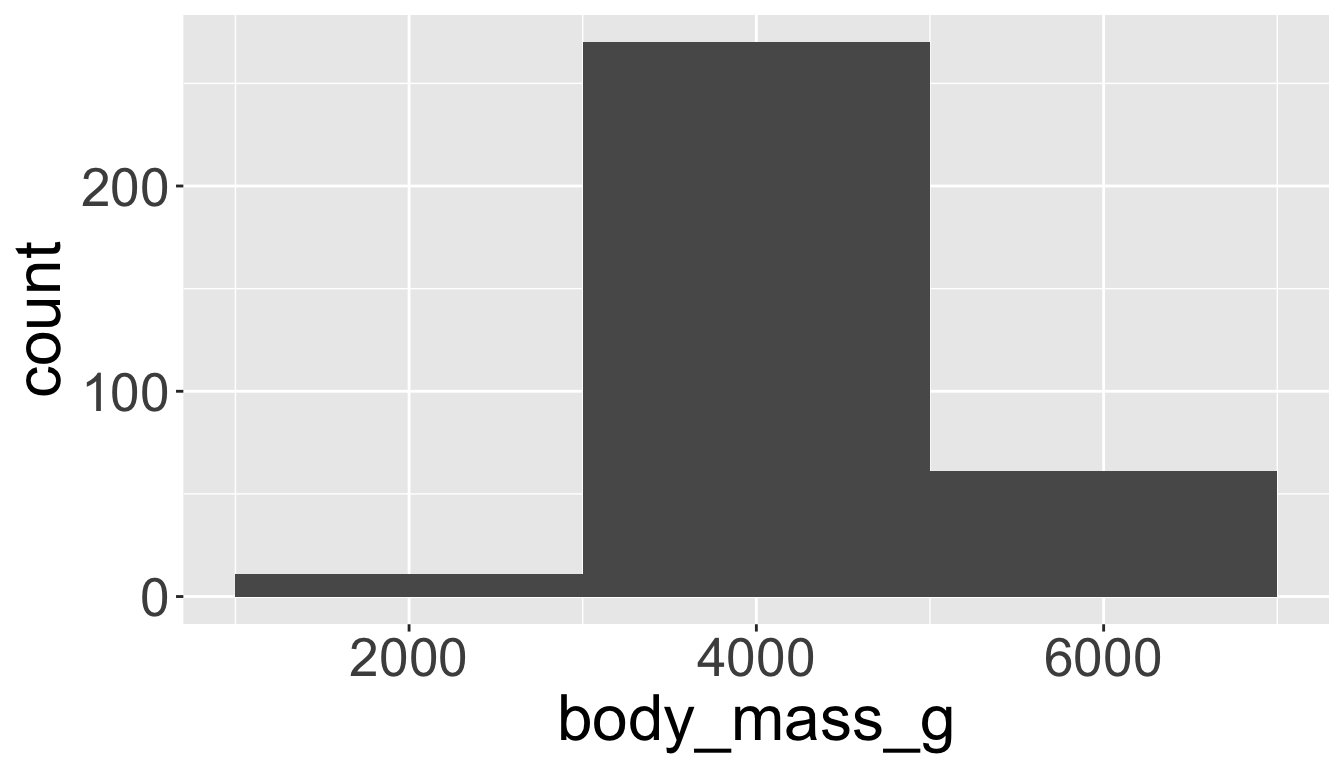
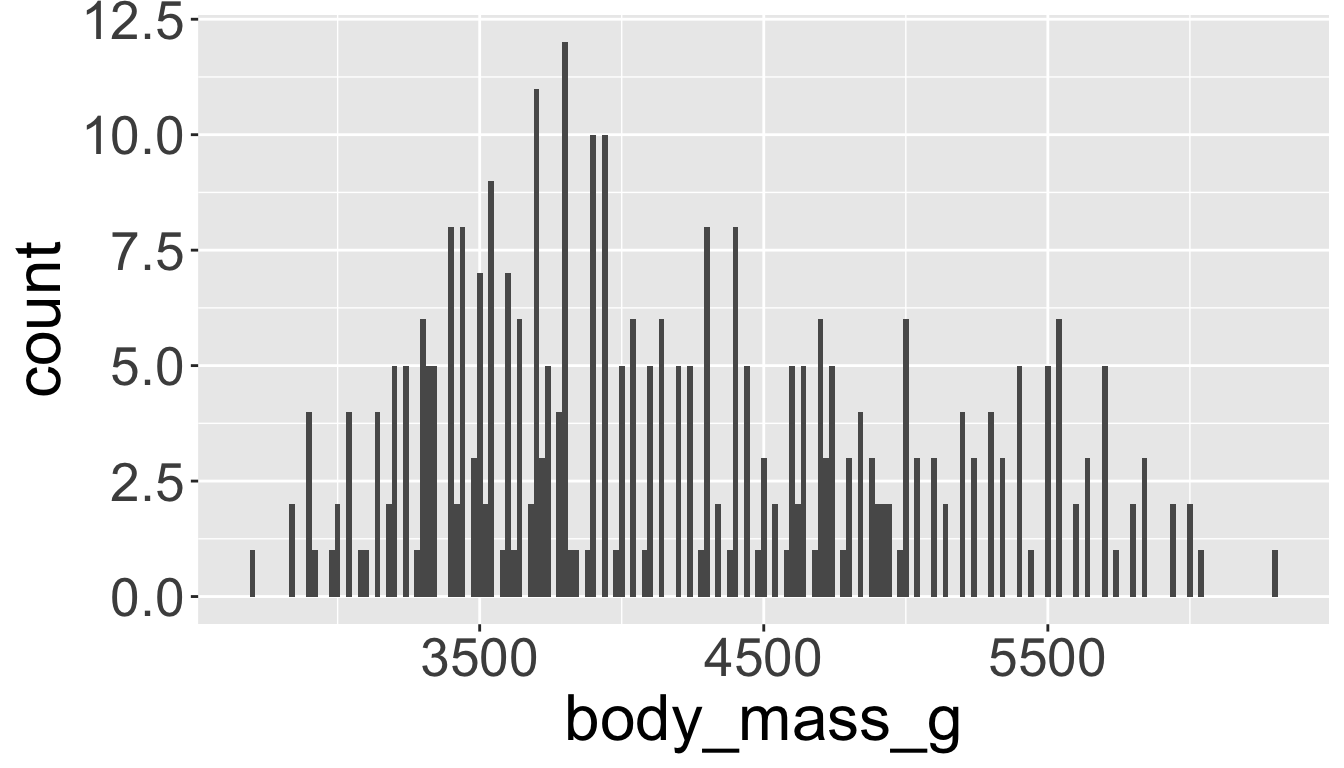
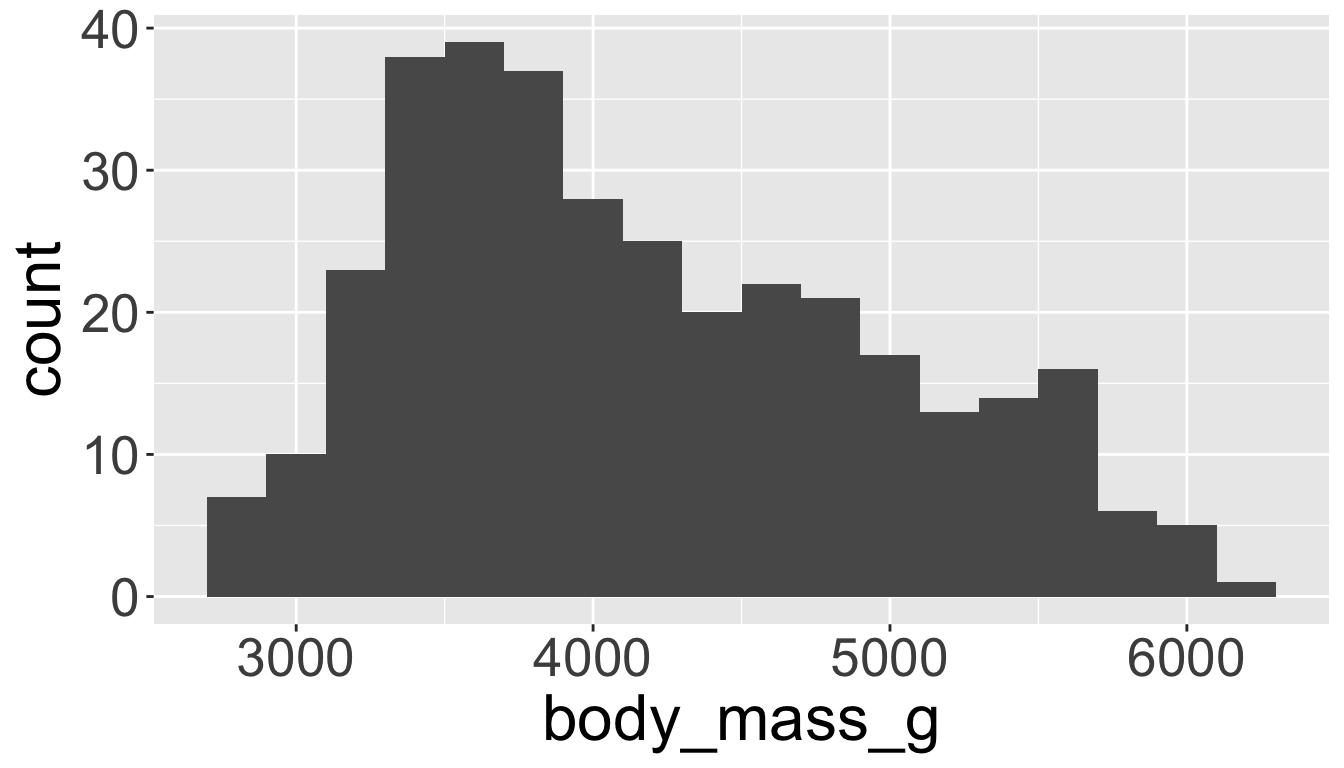
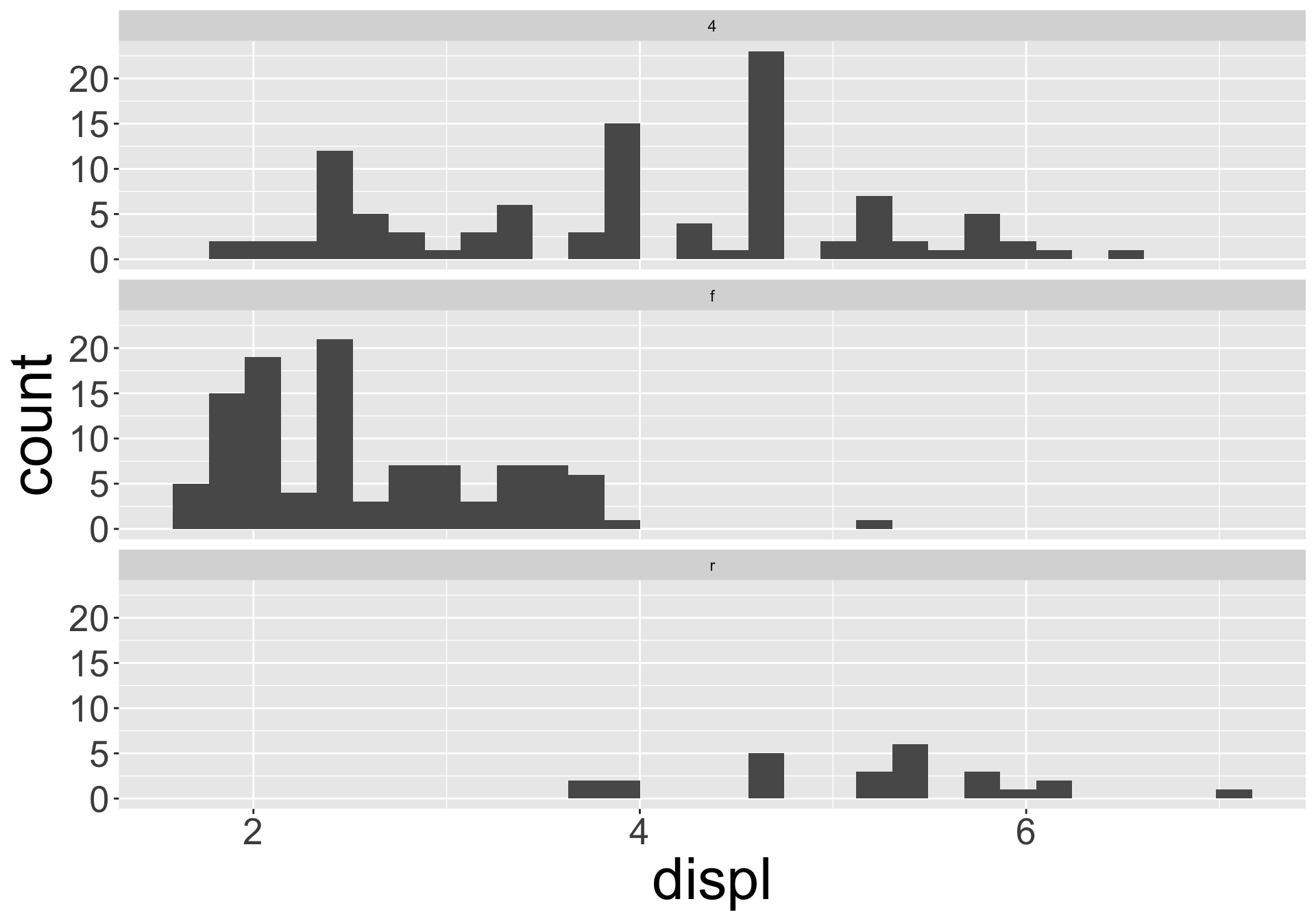
Visualizing distributions
You will likely need to spend time tuning the binwidth parameter
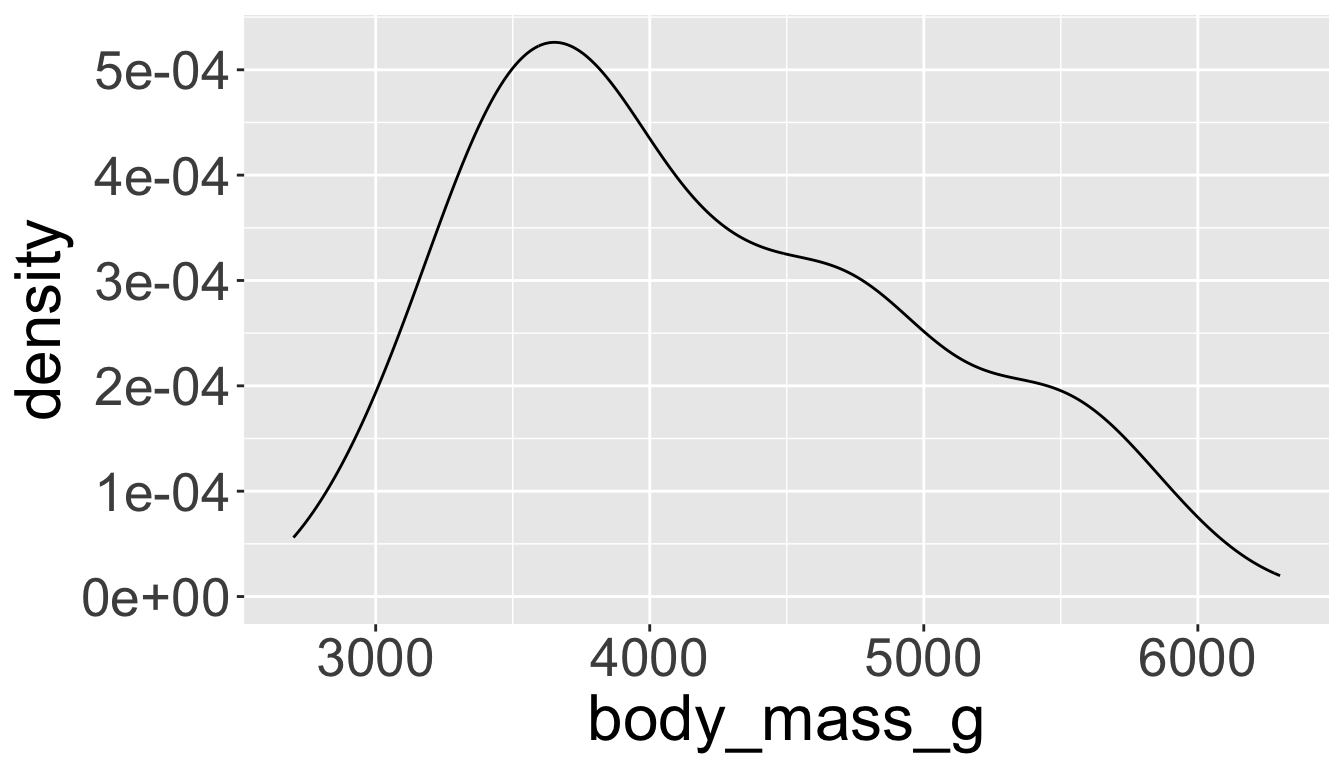
Visualizing distributions
- A smoothed out version of histogram which is supposed to approximate a probability density function
Visualizing distributions
- Let’s check the difference between setting
color =vsfill =withgeom_bar:
Visualizing distributions
- Let’s check the difference between setting
color =vsfill =withgeom_bar:
Visualizing distributions
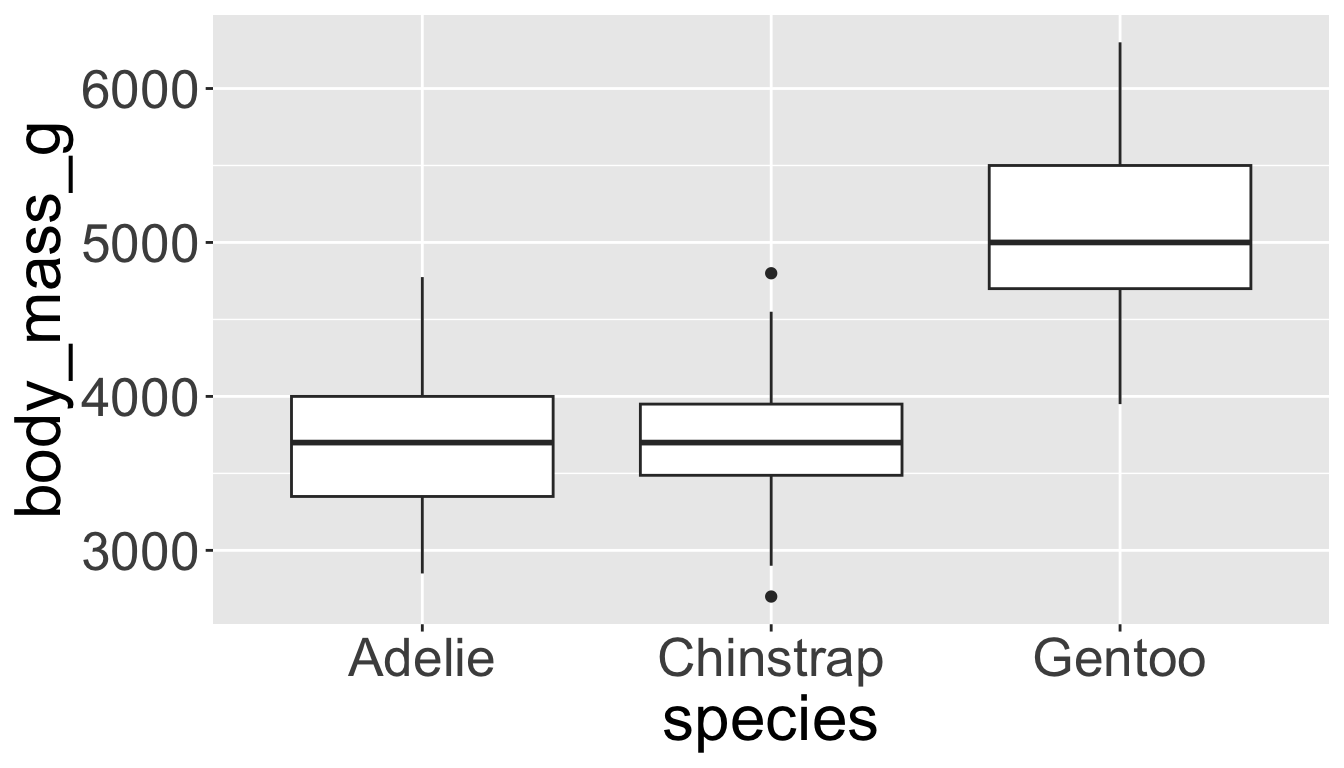
- Box plots allow for visualizing the spread of a distribution
- Makes it easy to see 25th percentile, median, 75th percentile, and outliers (>1.5*IQR from 25th or 75th percentile)
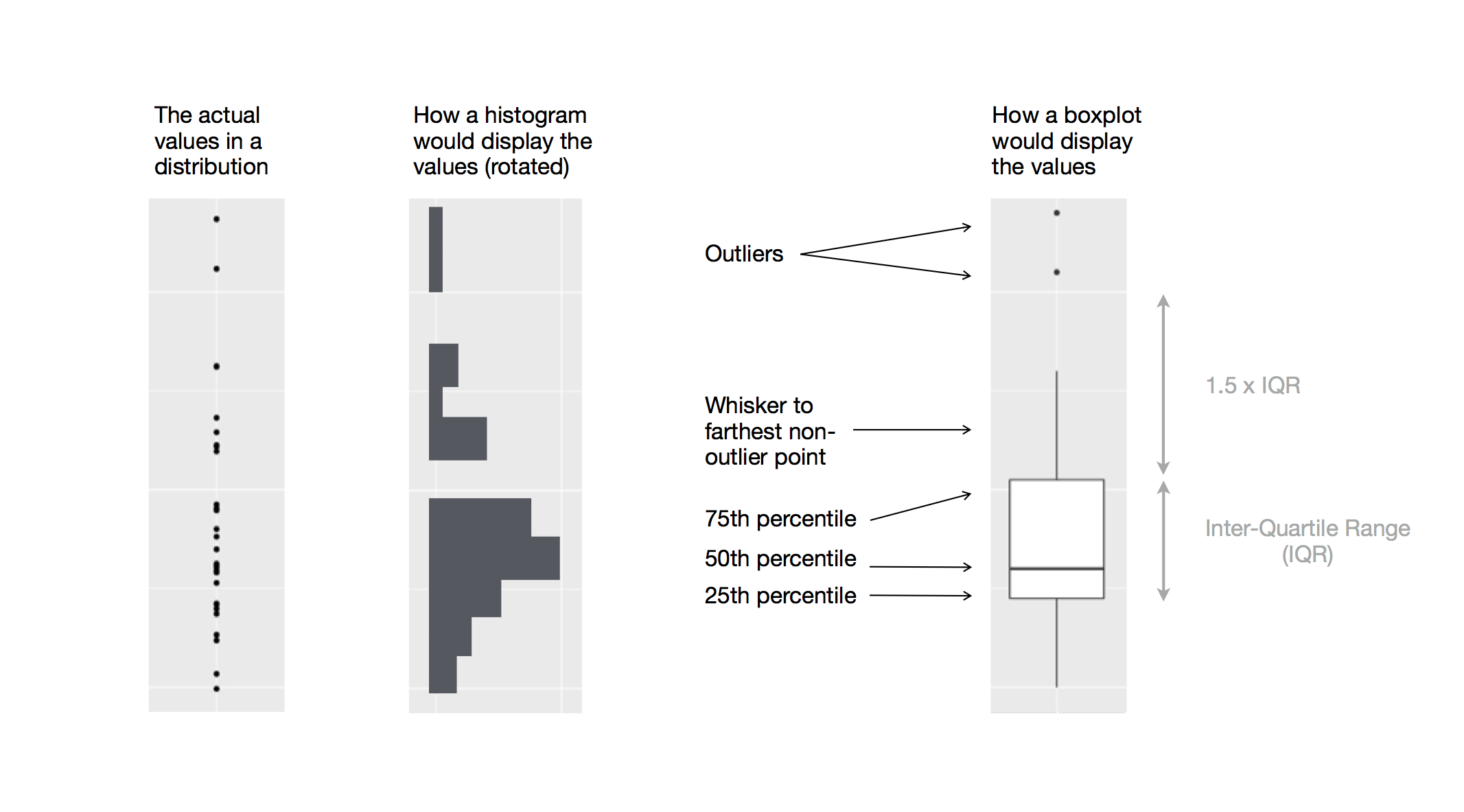
Visualizing distributions
Let’s see distribution of body mass by species…
Playing with visual parameters
Use alpha to add transparency
alphais a number between 0 and 1; 0 = transparent, 1 = opaque
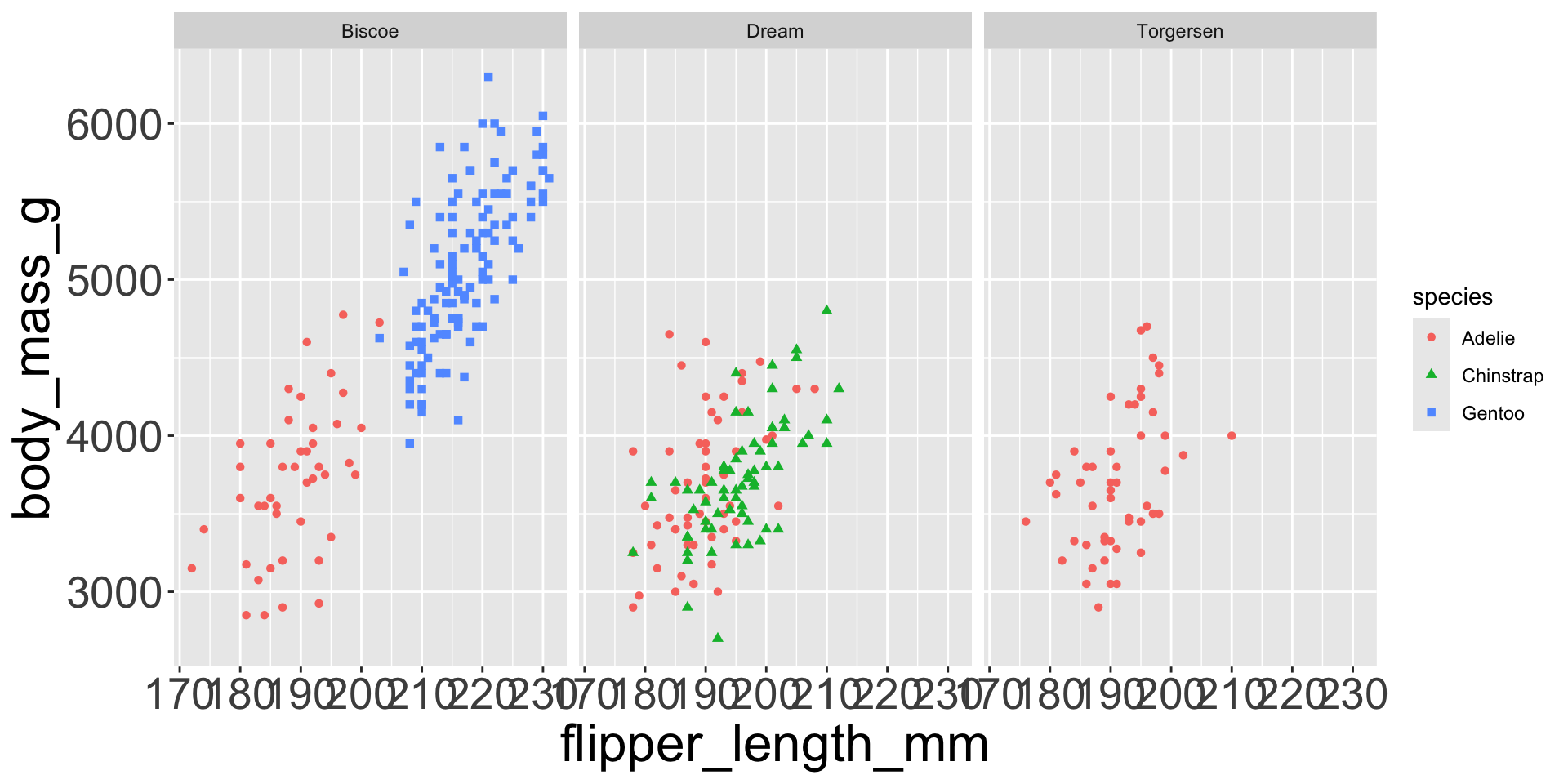
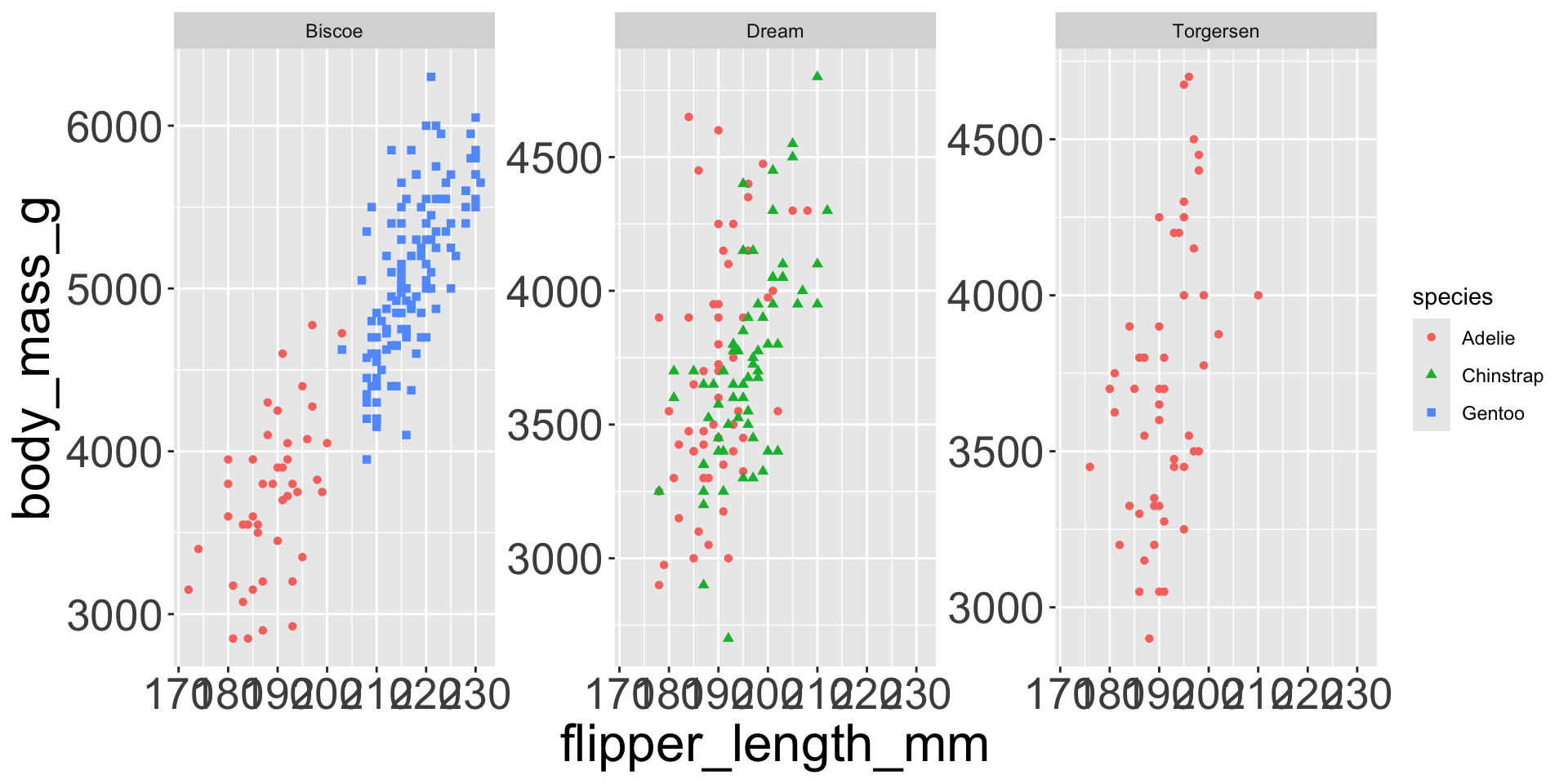
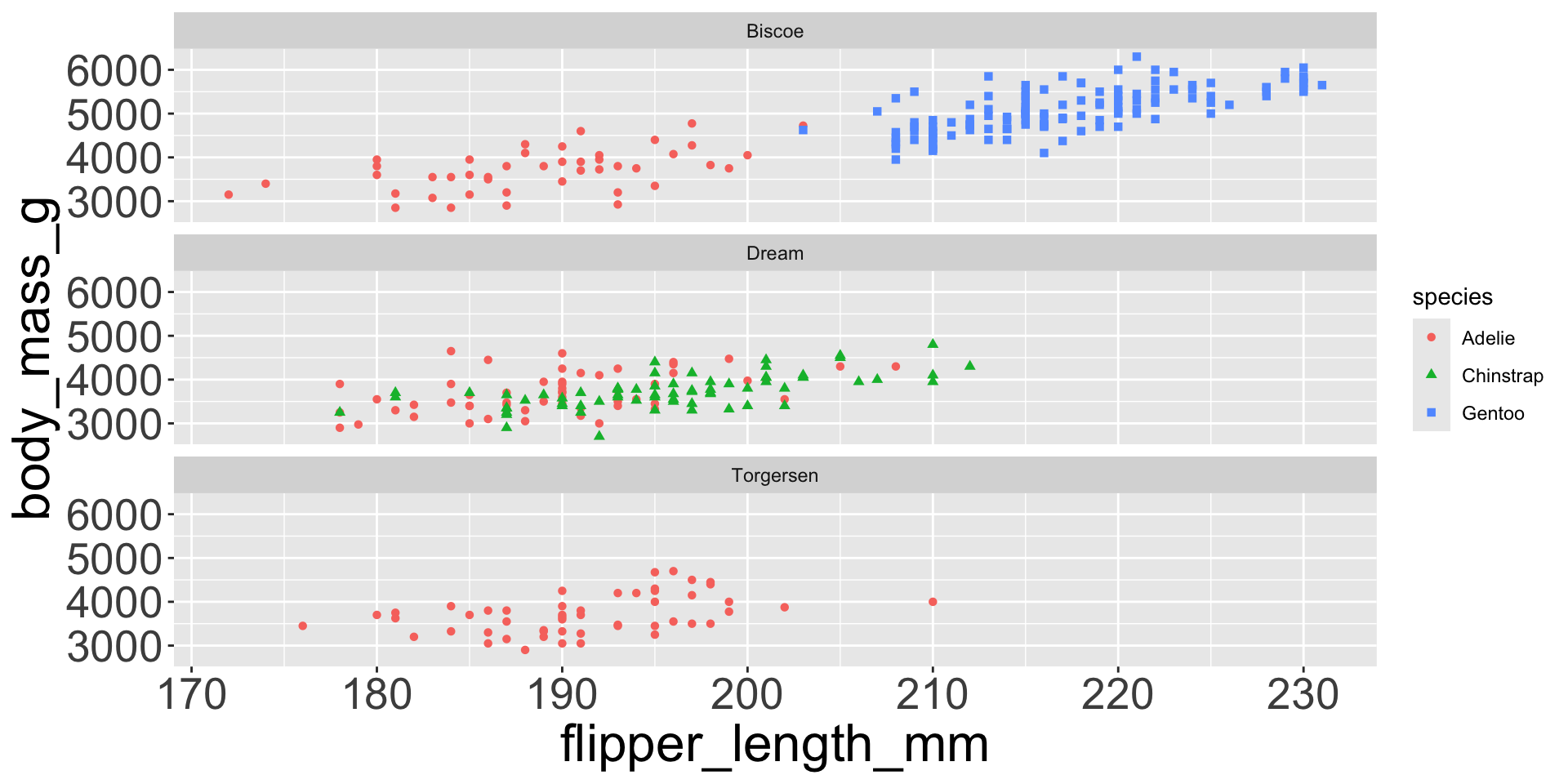
Multiple numerical variables
Already saw how to use scatter plots to visualize two numeric variables
Multiple numerical variables
Too many aesthetic changes (shape, color, fill, size, etc) can clutter plots
Multiple numerical variables
Too many aesthetic changes (shape, color, fill, size, etc) can clutter plots
Multiple numerical variables
Too many aesthetic changes (shape, color, fill, size, etc) can clutter plots
Multiple numerical variables
Too many aesthetic changes (shape, color, fill, size, etc) can clutter plots
Multiple numerical variables
Too many aesthetic changes (shape, color, fill, size, etc) can clutter plots
Multiple numerical variables
Too many aesthetic changes (shape, color, fill, size, etc) can clutter plots
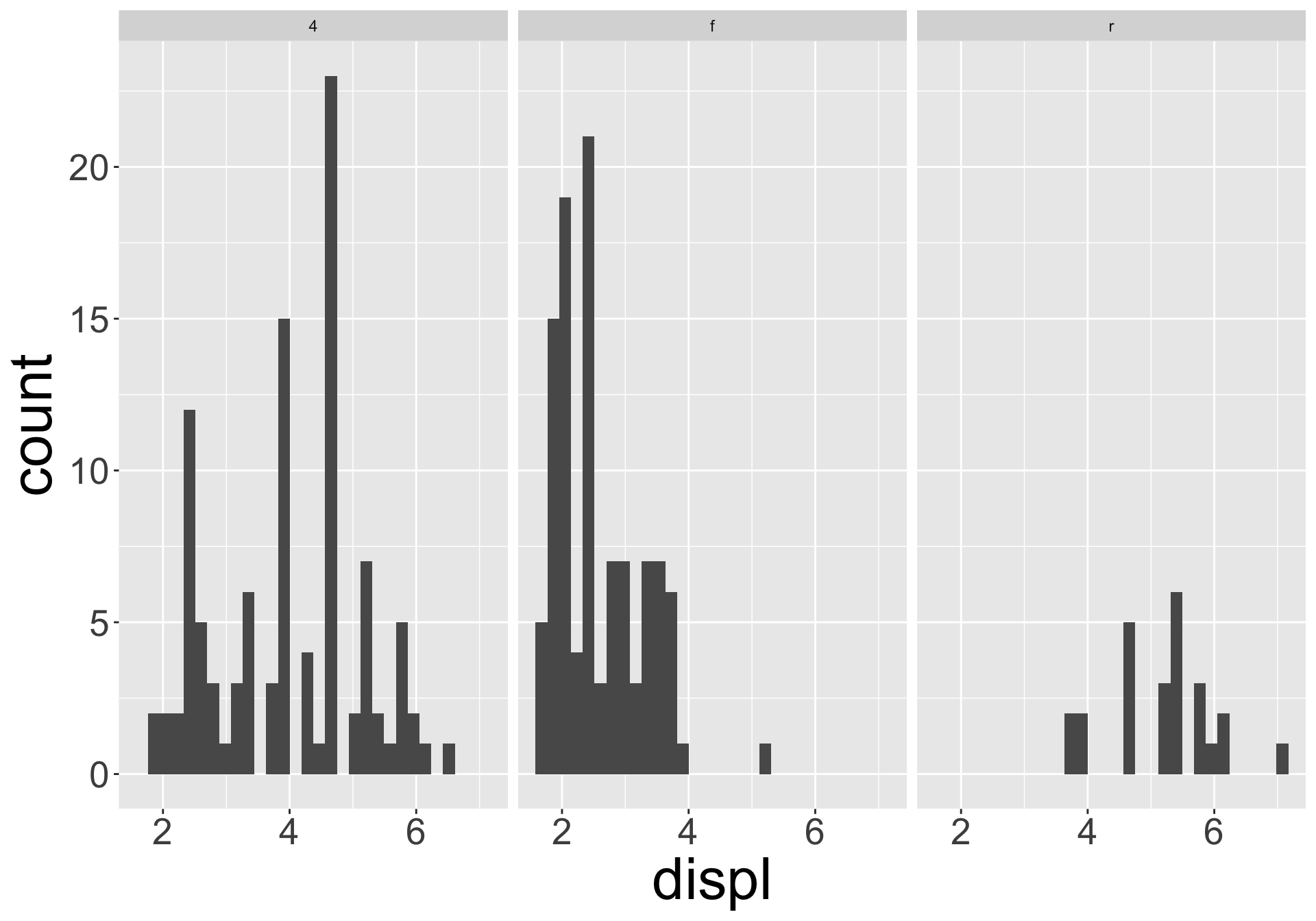
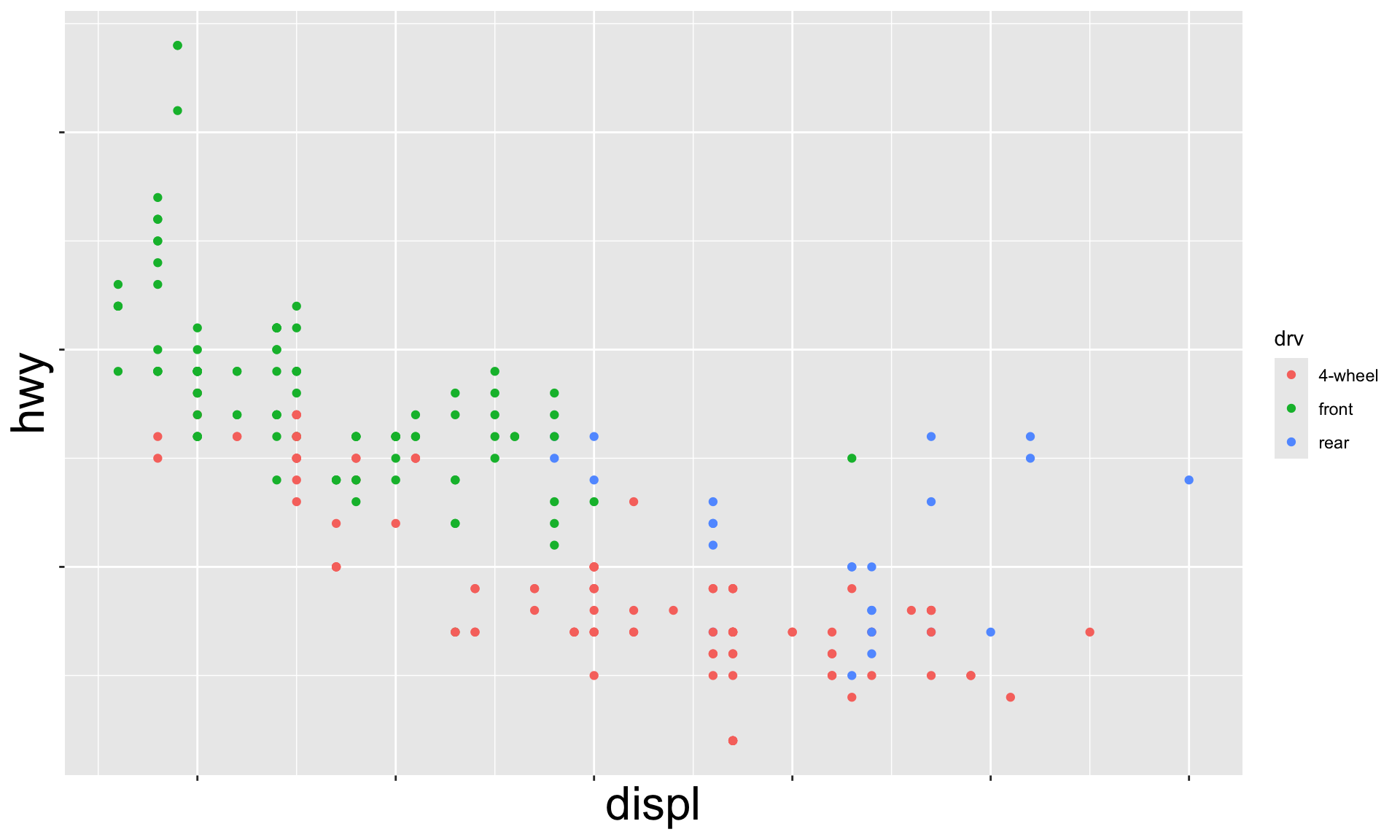
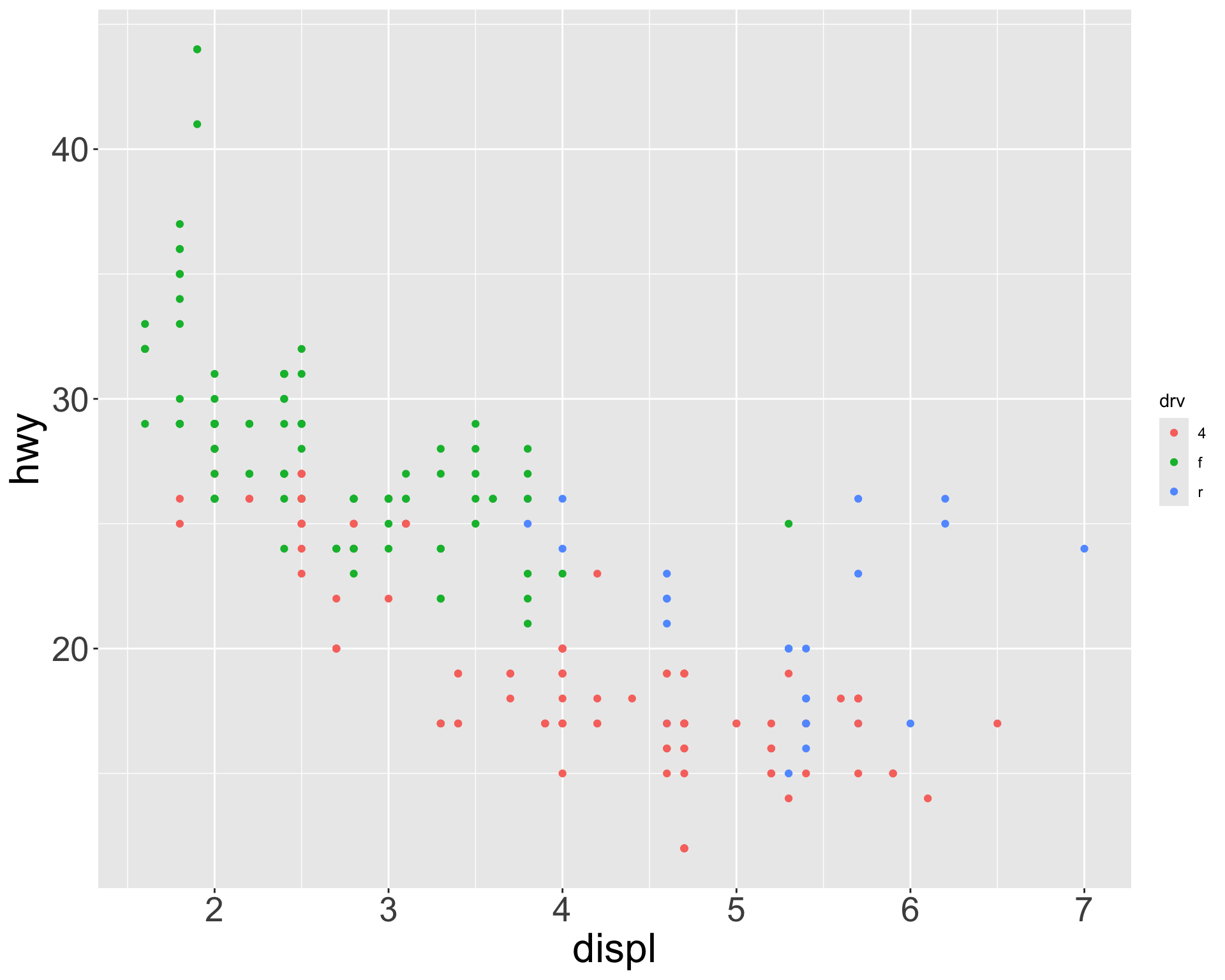
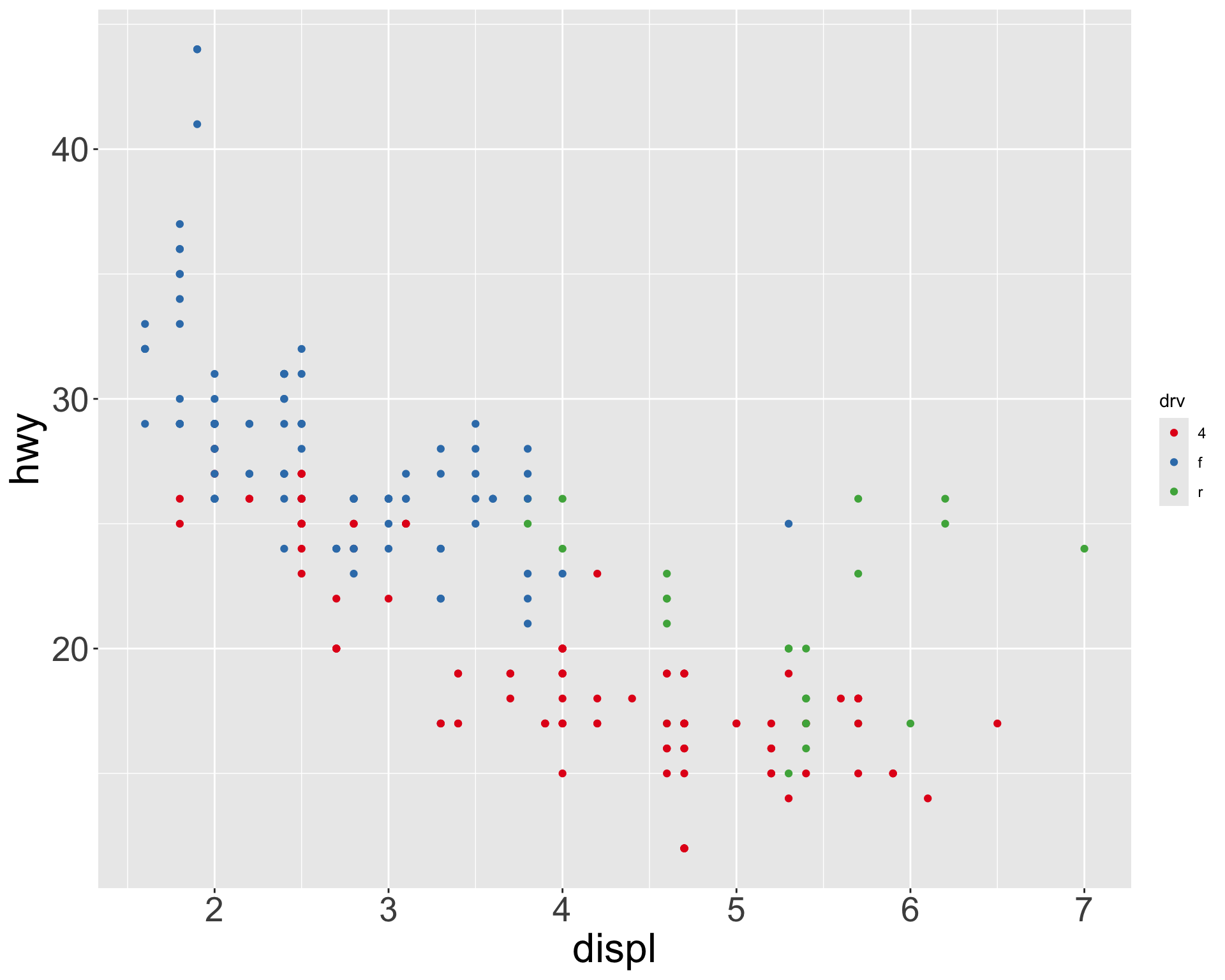
Multiple numerical variables
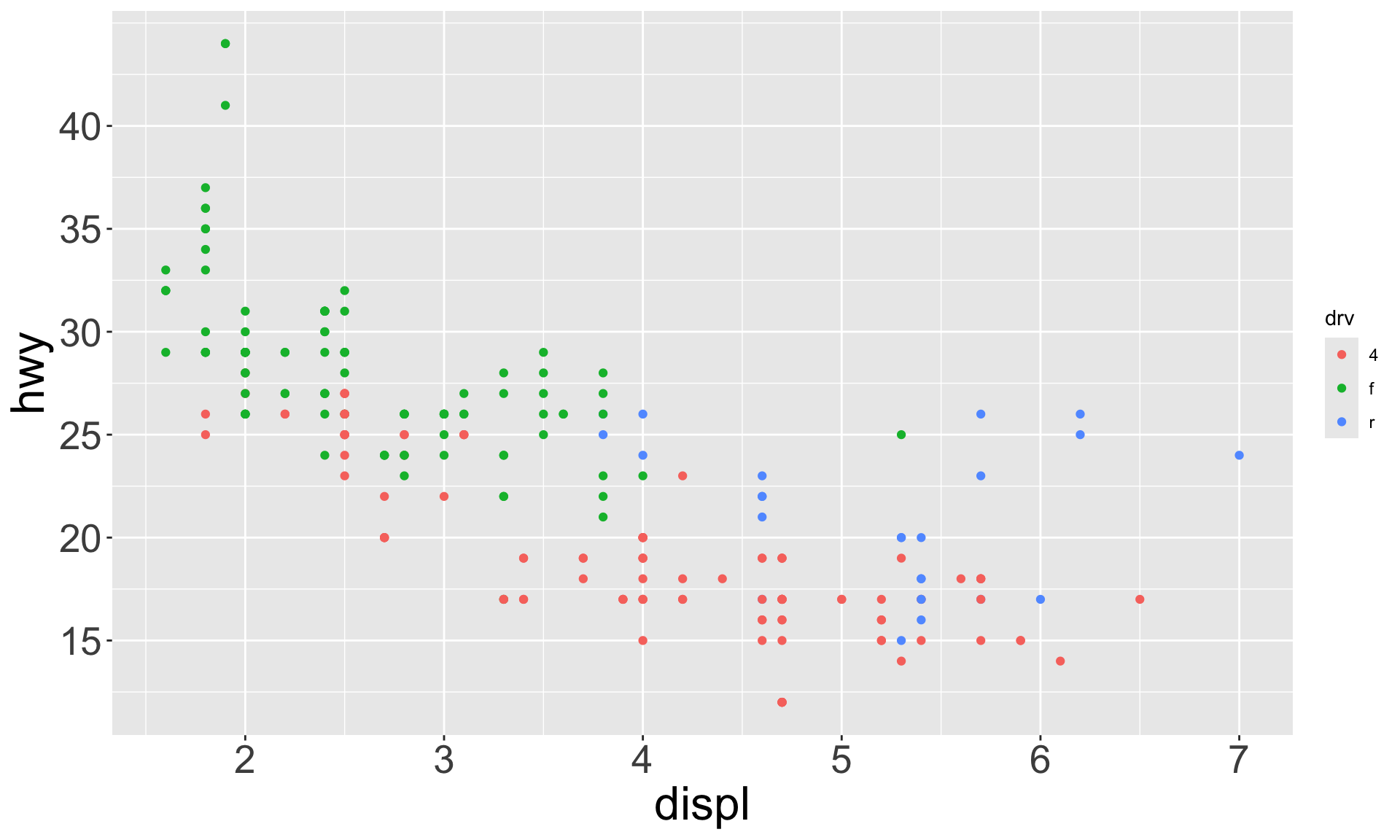
Which of the following makes it easier to compare engine size (displ) across cars with different drive trains?
Can change order of levels in factors
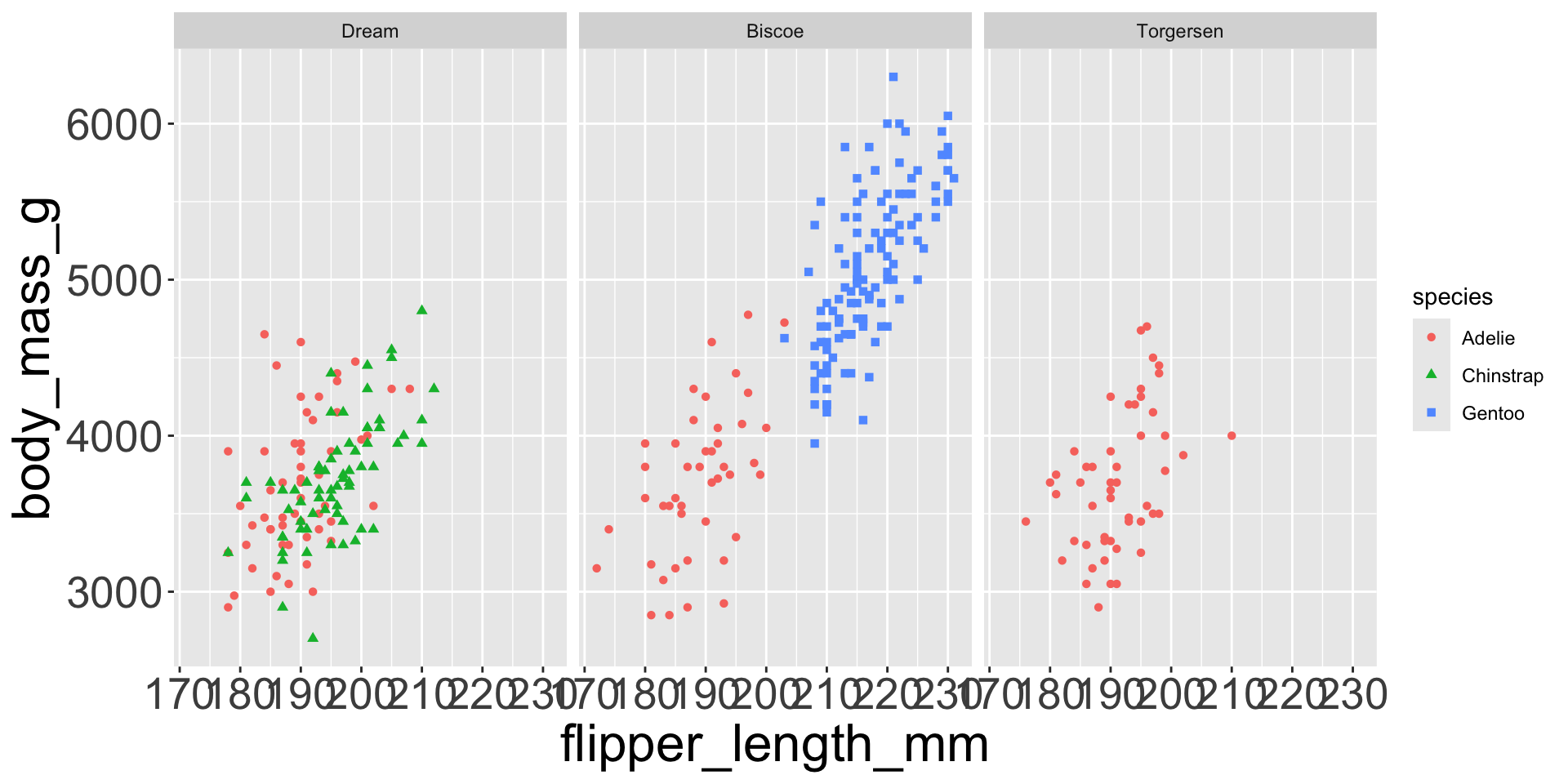
Shapes can be difficult to distinguish
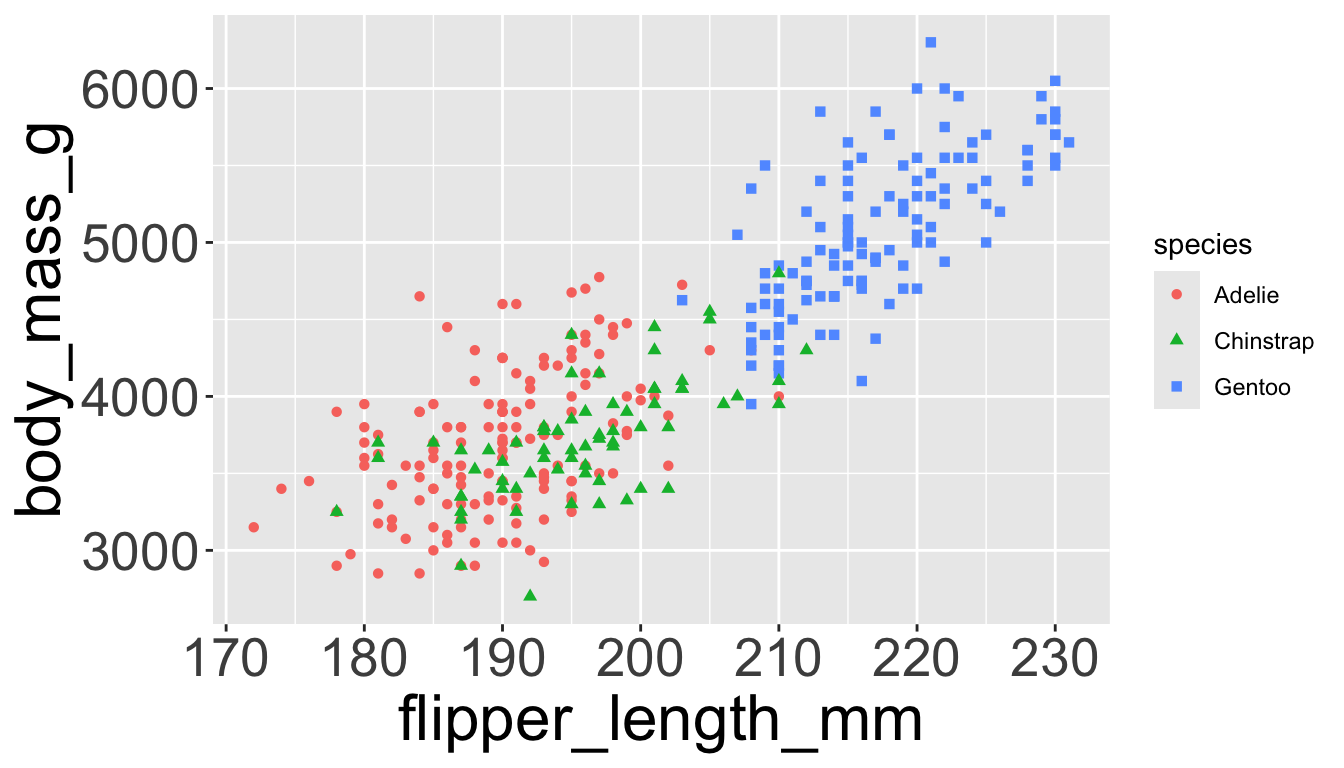
Sometimes easier to read by replacing shape with first letter
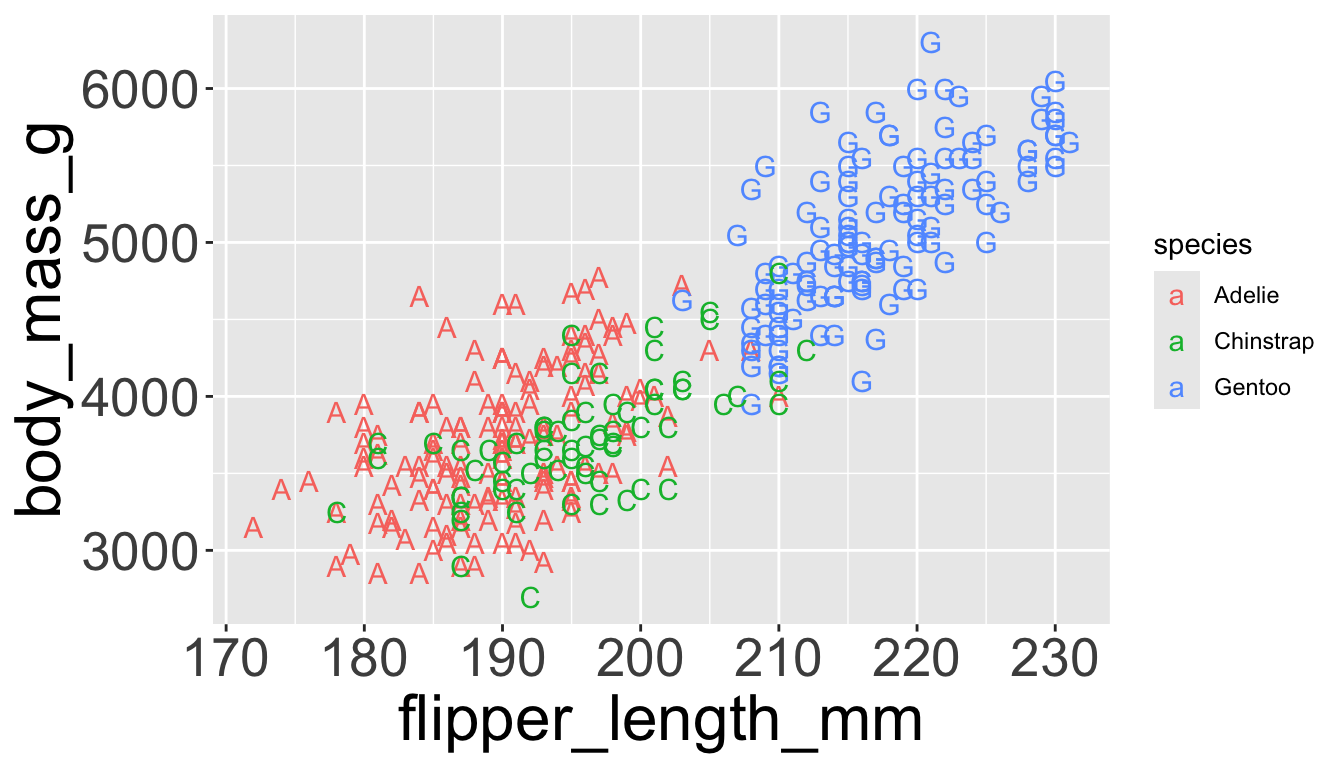
Might allow you to remove the legend
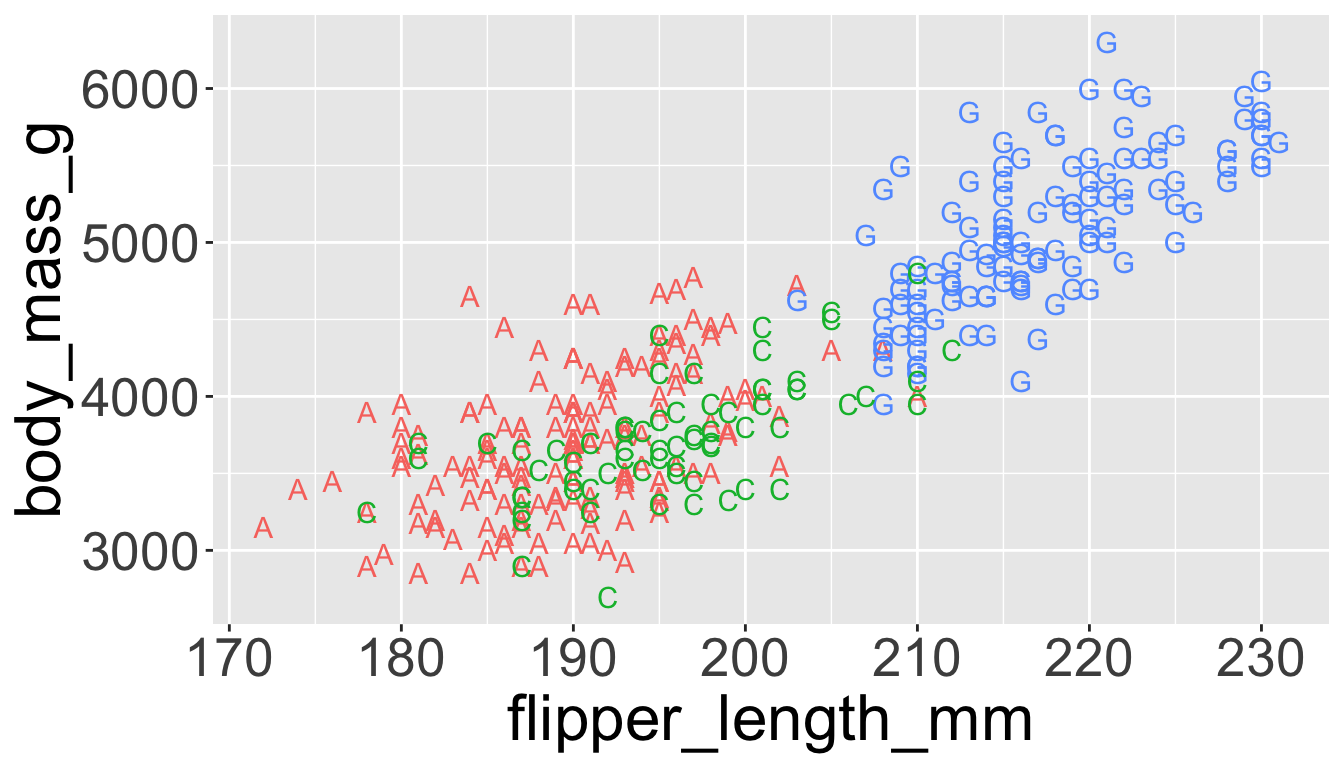
Saving plots
Once you’ve made a plot, you can save using ggsave()
- Either can save whatever plot you made last:
- Or you can save the plot object and save that
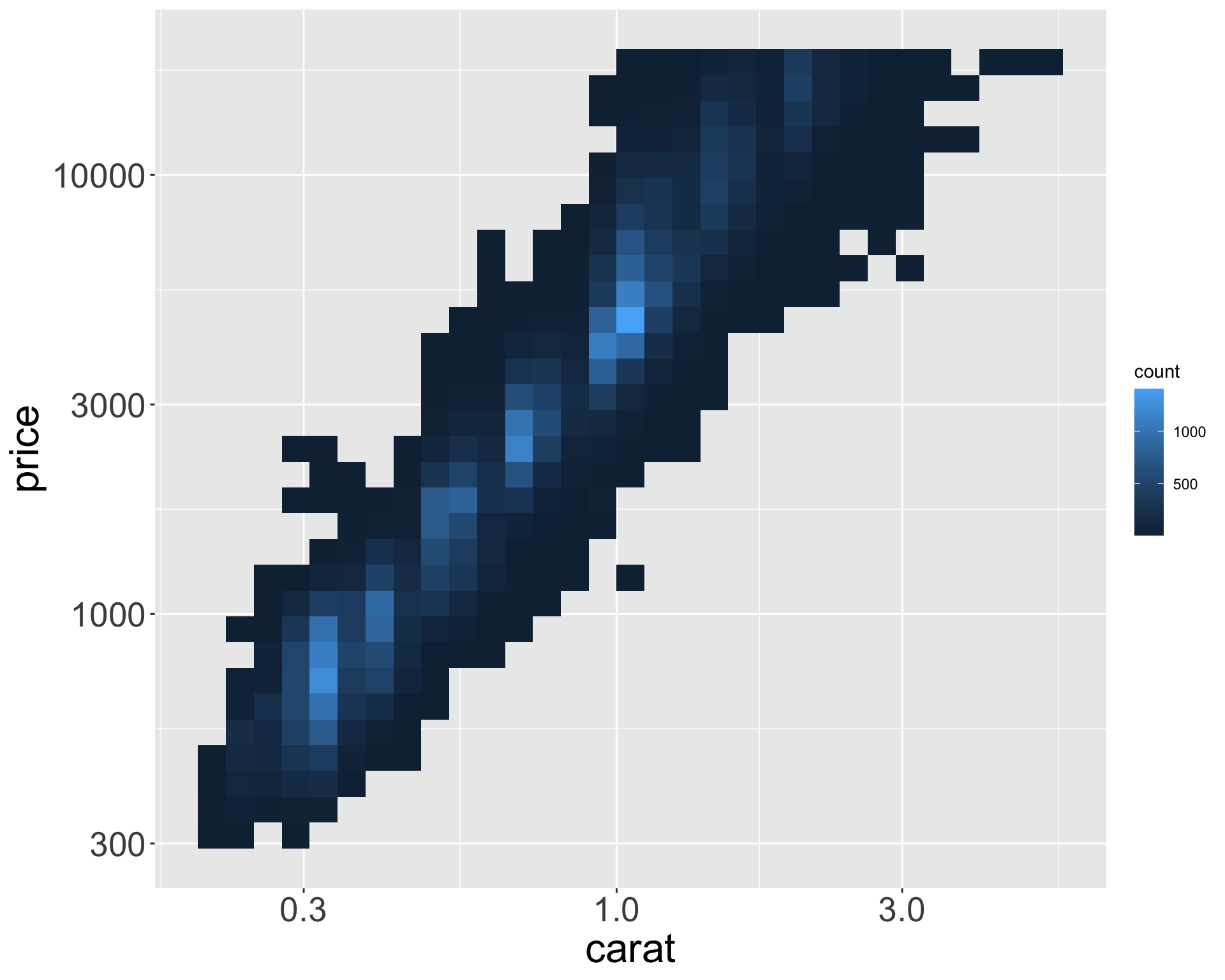
Exploring the diamonds data set
Diamonds data set
Consider the diamonds dataset in ggplot2
- Contains the prices and other attributes of almost 54,000 diamonds.
# A tibble: 53,940 × 10
carat cut color clarity depth table price x y z
<dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
4 0.29 Premium I VS2 62.4 58 334 4.2 4.23 2.63
5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
# ℹ 53,930 more rowsGeometric objects
How are these two plots similar?
- Both plots contain the same x and y variables; both plots describe same data.
- Each plot uses a different geometric object,
geom, to represent the data.
Geometric objects
- Can utilize multiple
geoms together to elucidate relationship
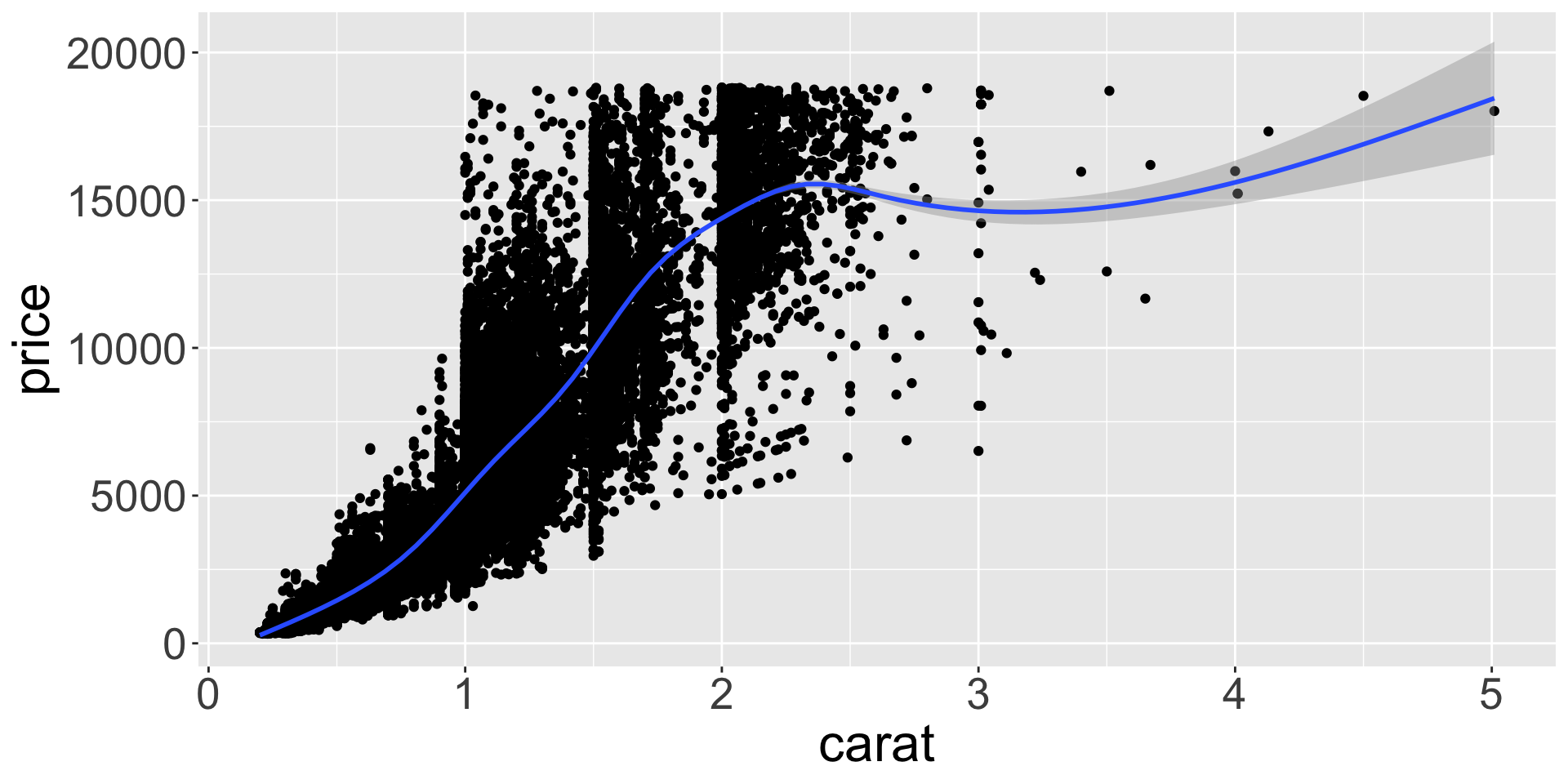
Statistical transformations
Let’s create a bar chart across cuts:
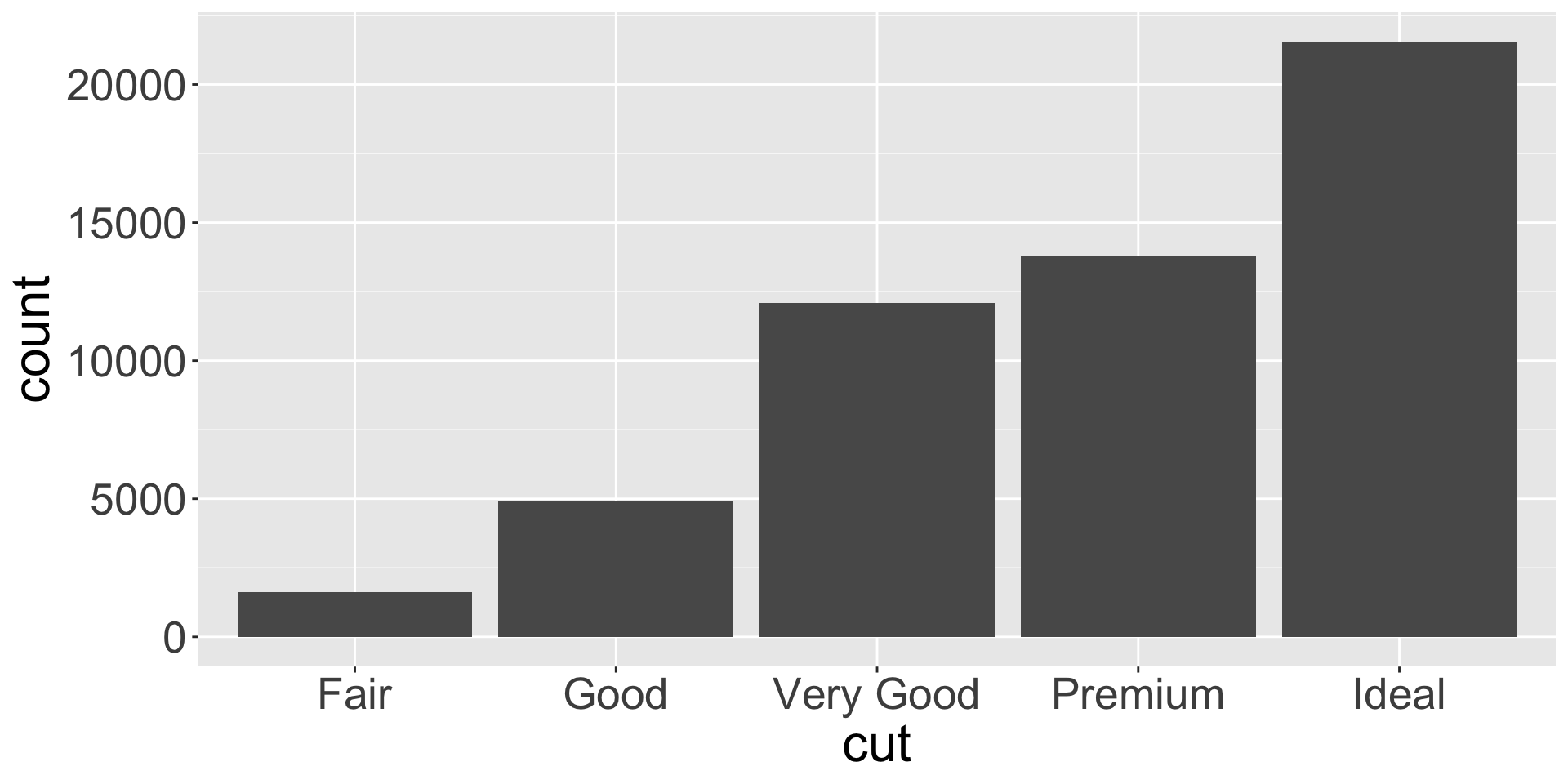
countis not a variable in diamonds, so how is it creating this?
Statistical transformations
Examples
- Bar charts, histograms, and frequency polygons bin your data, then plot bin counts (the number of points that fall in each bin).
- Smoothers fit a model to your data and then plot predictions from the model.
- Boxplots compute the five-number summary of the distribution and then display that summary as a specially formatted box.
Statistical transformations
The algorithm used to calculate new values for a graph is called a stat.
- (stat is short for statistical transformation)
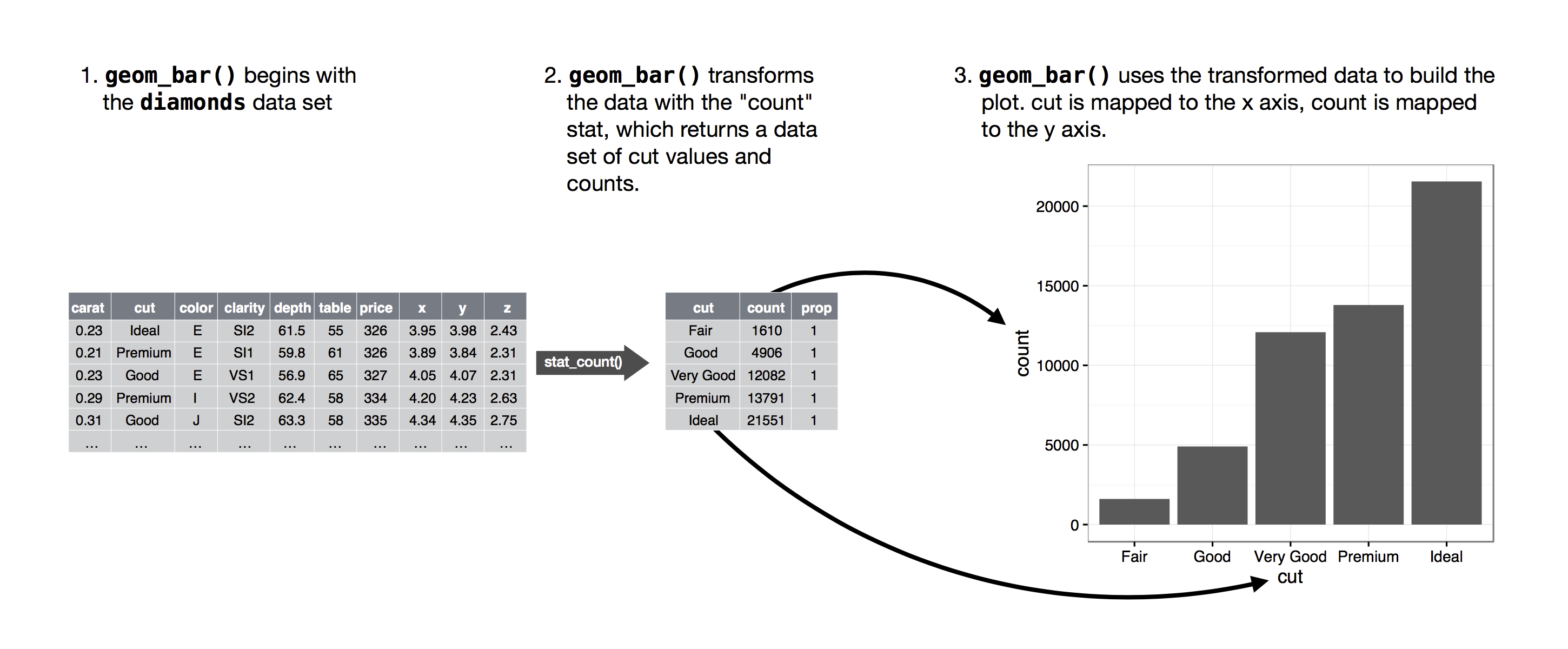
Figure 2
Exploring variables
Two question categories:
- What type of variation occurs within each variable?
- What type of covariation occurs between 2+ variables?
Variation
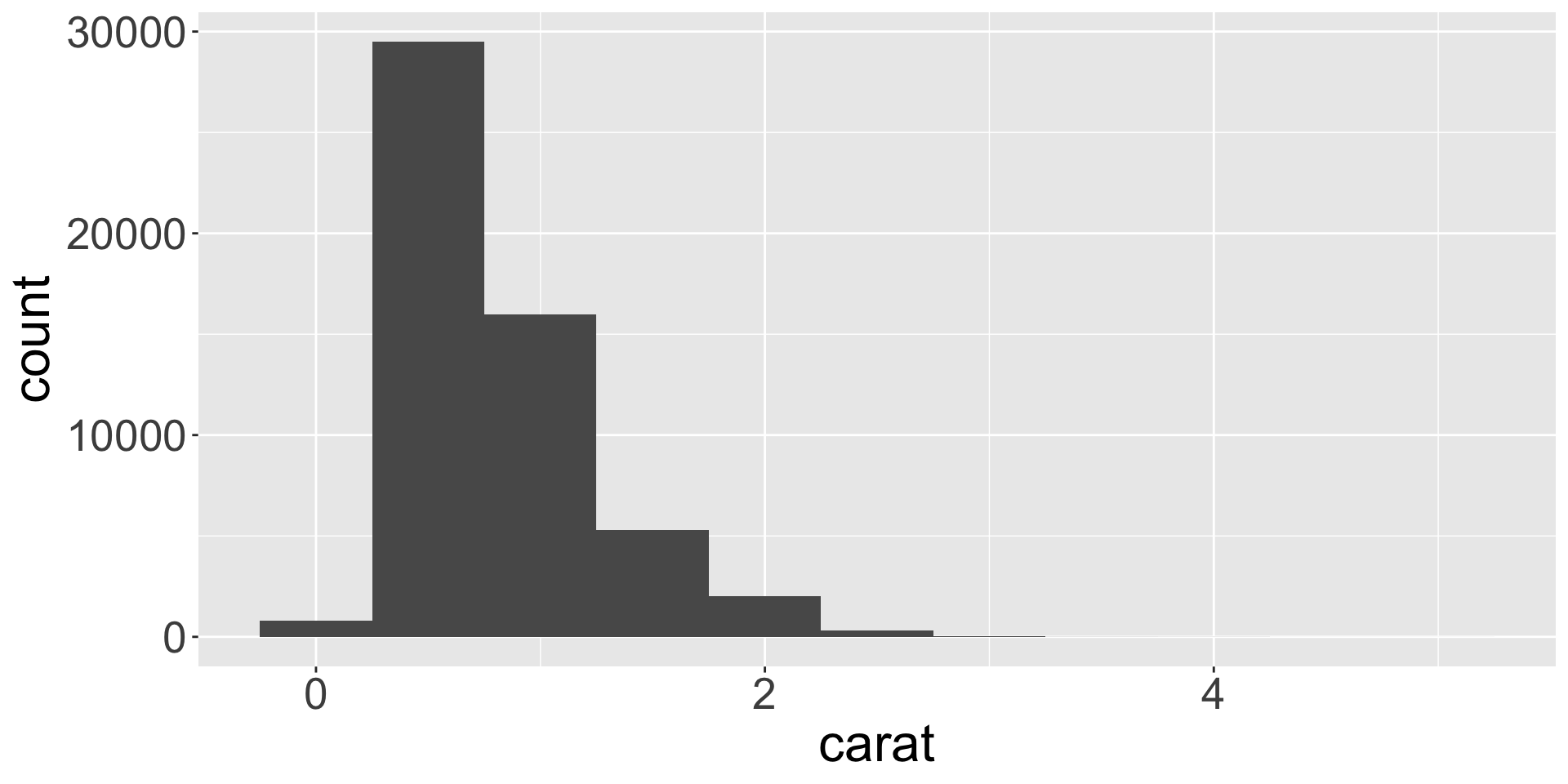
Variation is tendency for values of a variable to change from measurement to measurement
- Can be due to measurement error (e.g., measuring height with different rulers) or due to within-group variation (different people have different heights)
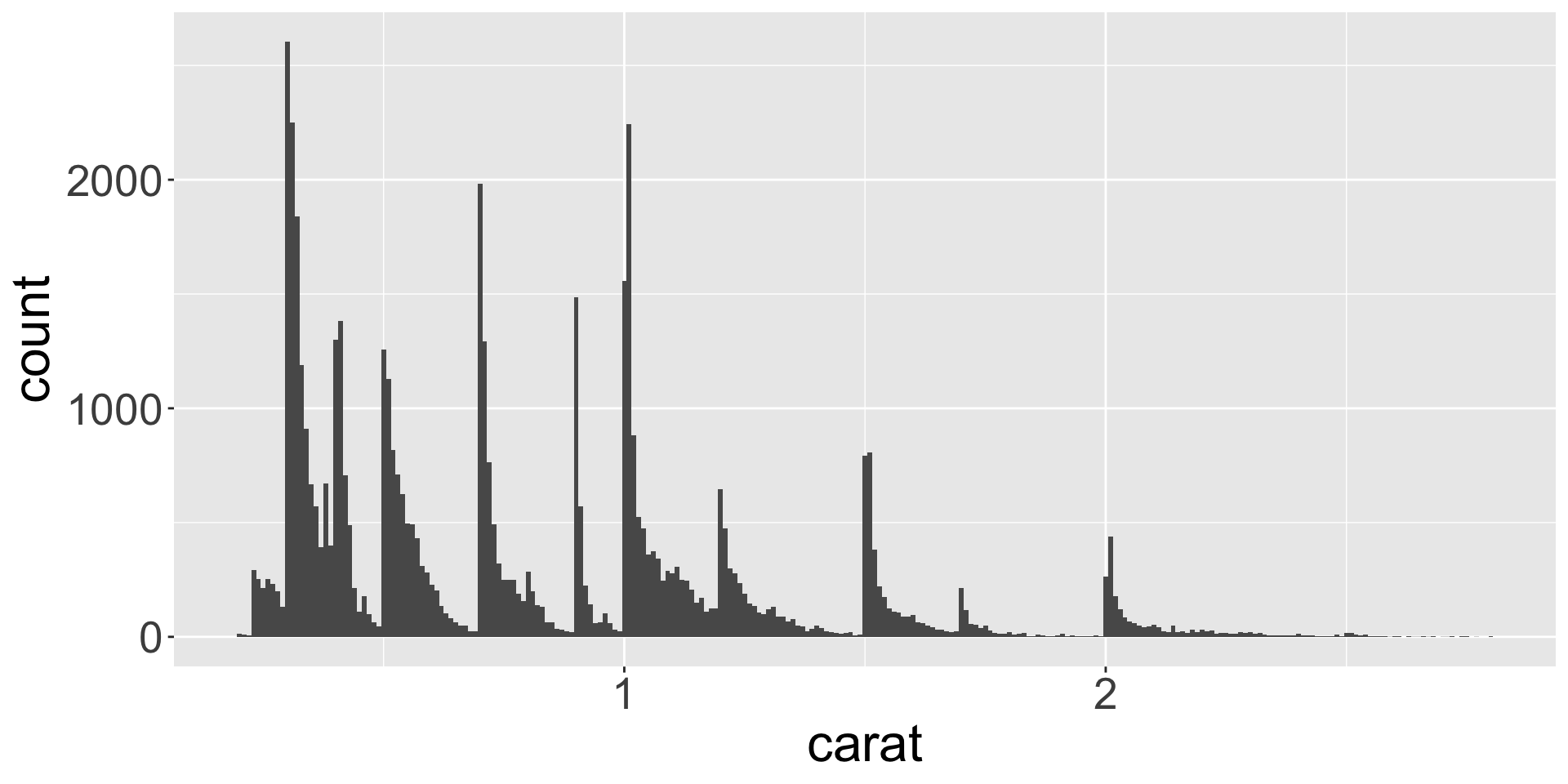
- Let’s explore the distribution of weights (
carat) of the ~50k diamonds fromdiamondsdataset. caratis numerical, can use histogram:
Typical values
- In bar charts and histograms, tall bars = common values; no bars = values not seen
- Questions to ask yourself:
- Which values are the most common? Why?
- Which values are rare? Why? Does that match your expectations?
- Can you see any unusual patterns? What might explain them?
- Let’s look at distribution of weights of smaller diamonds.
Unusual values
If you encounter unusual values in dataset, and want to ignore them for rest of analysis, two options:
- Drop entire row with the strange values, e.g.
- Replacing unusual values with missing values
- Latter is more recommended behavior.
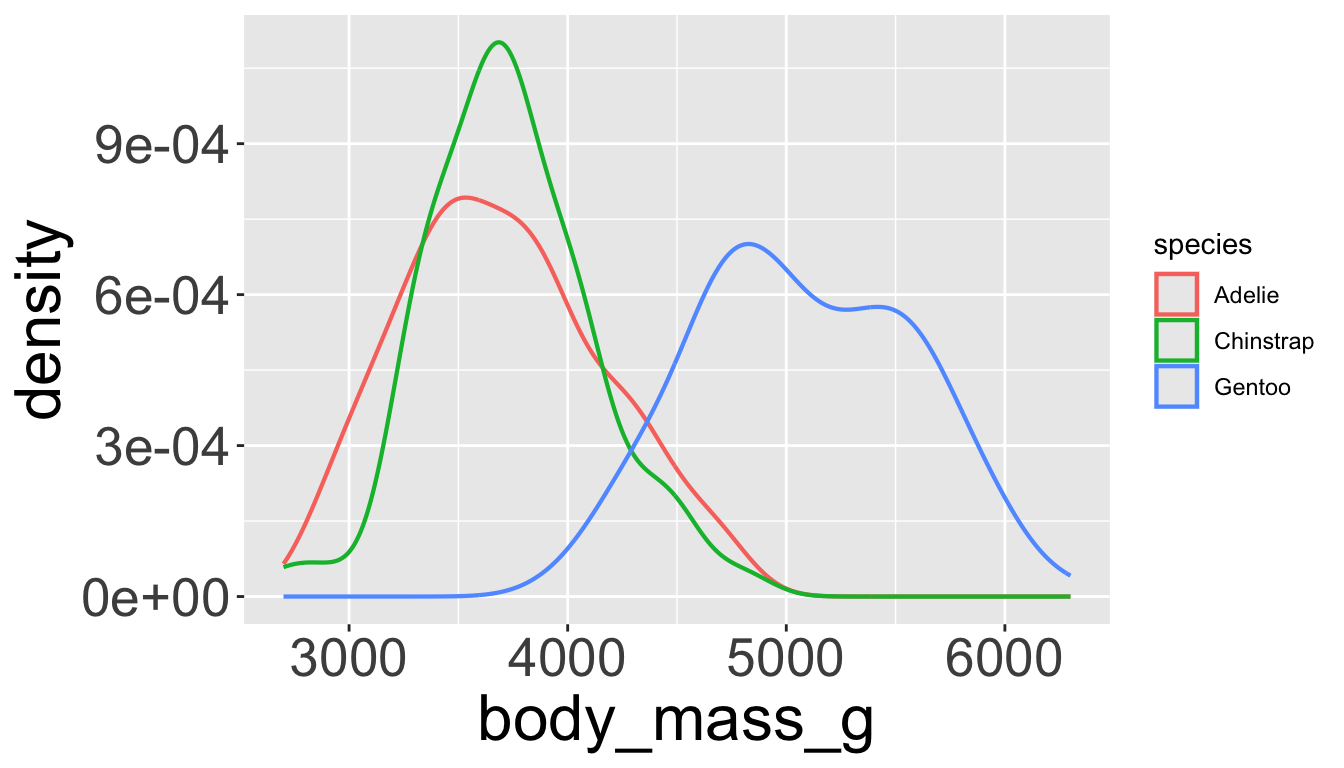
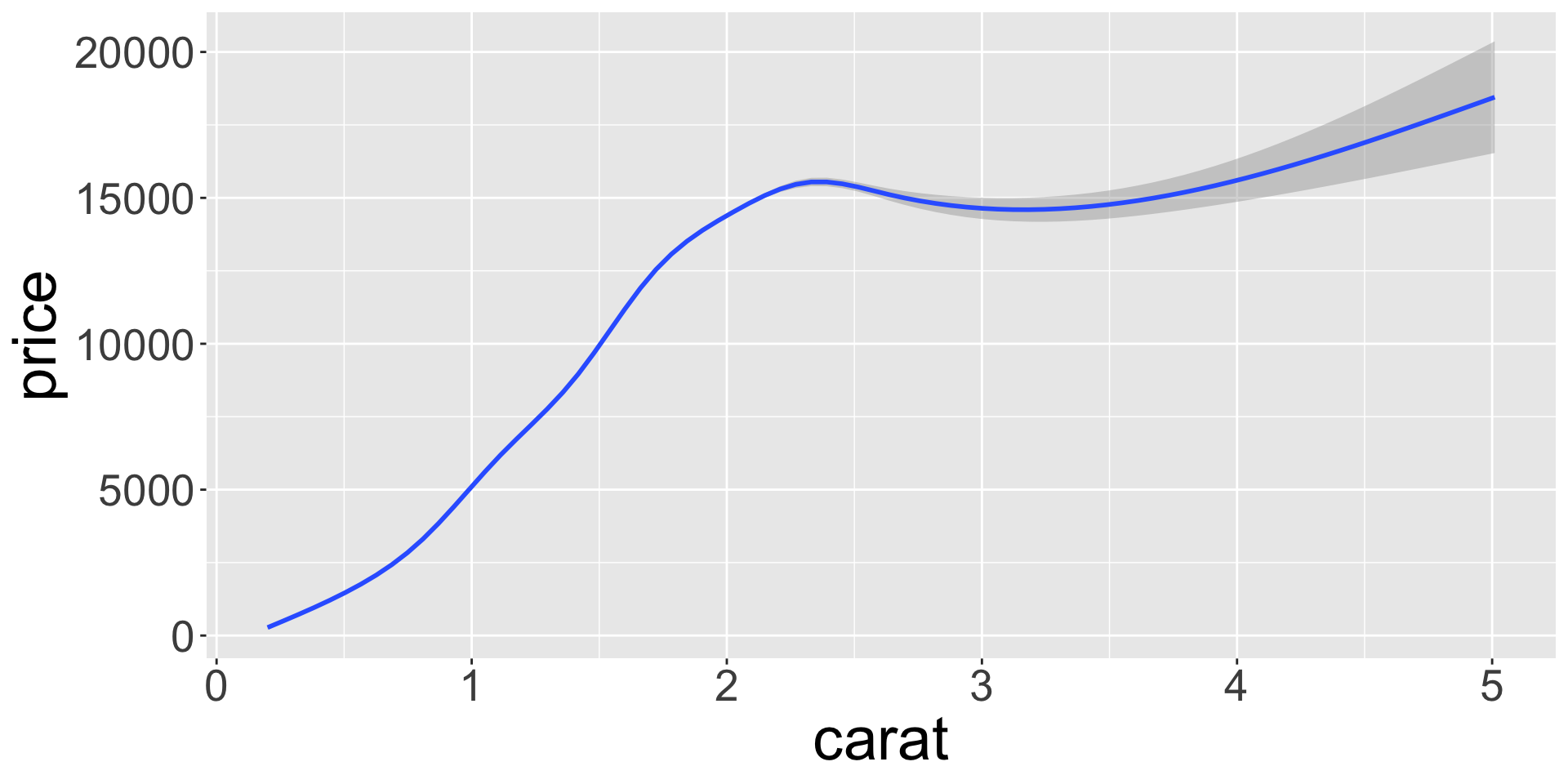
Covariation
… is tendency for values of 2+ variables to vary together in a related way
- How does price (numerical) of a diamond vary with quality (categorical)?
- Can use
geom_freqpoly()to show “frequency polygons” (similar to histogram)
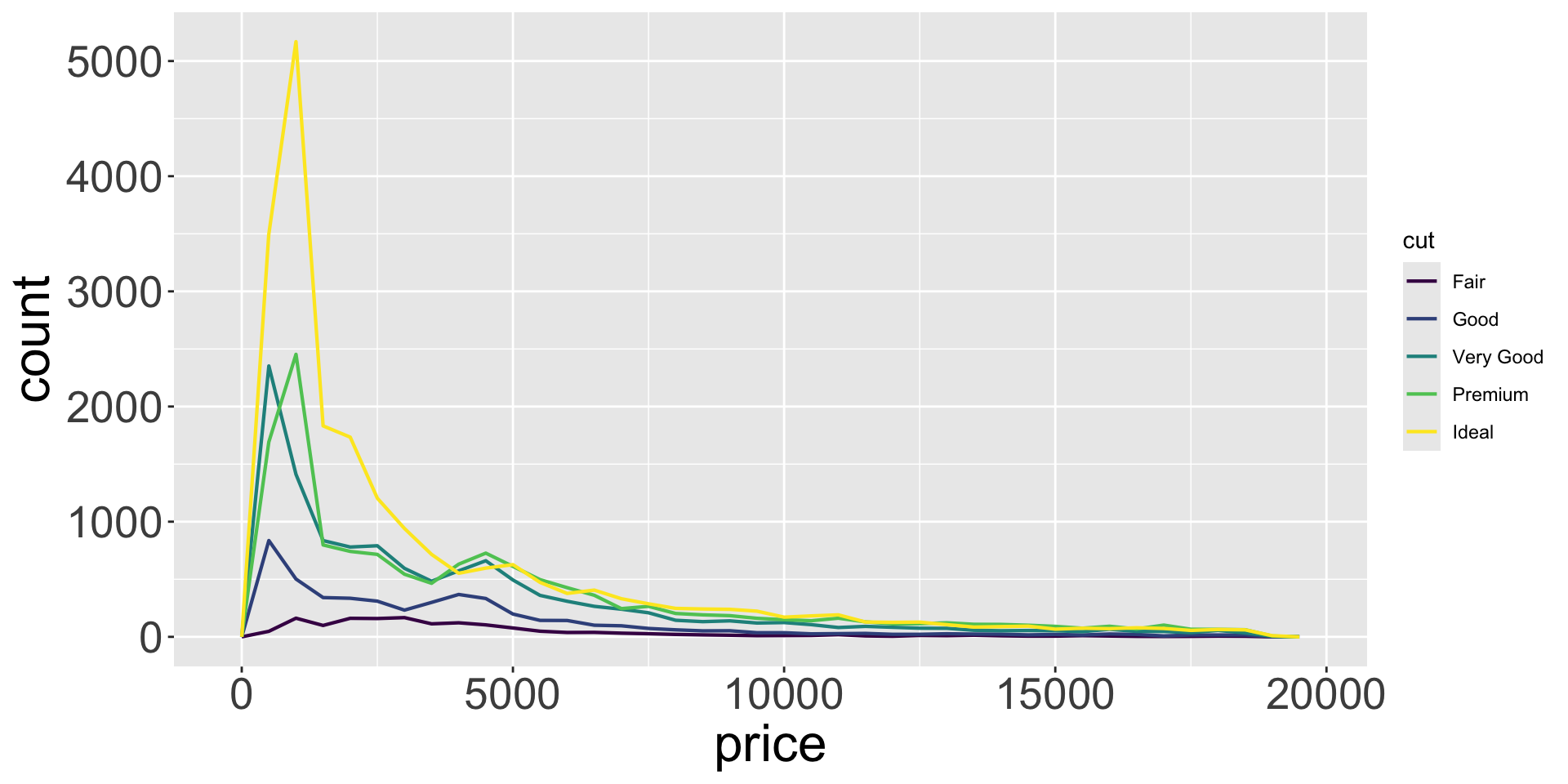
- Not super informative, since the height mainly reflects the count.
Covariation
- More useful to understand the density of the variable (count / total number)
- To do this, we can use
after_stat(density), which does this normalization
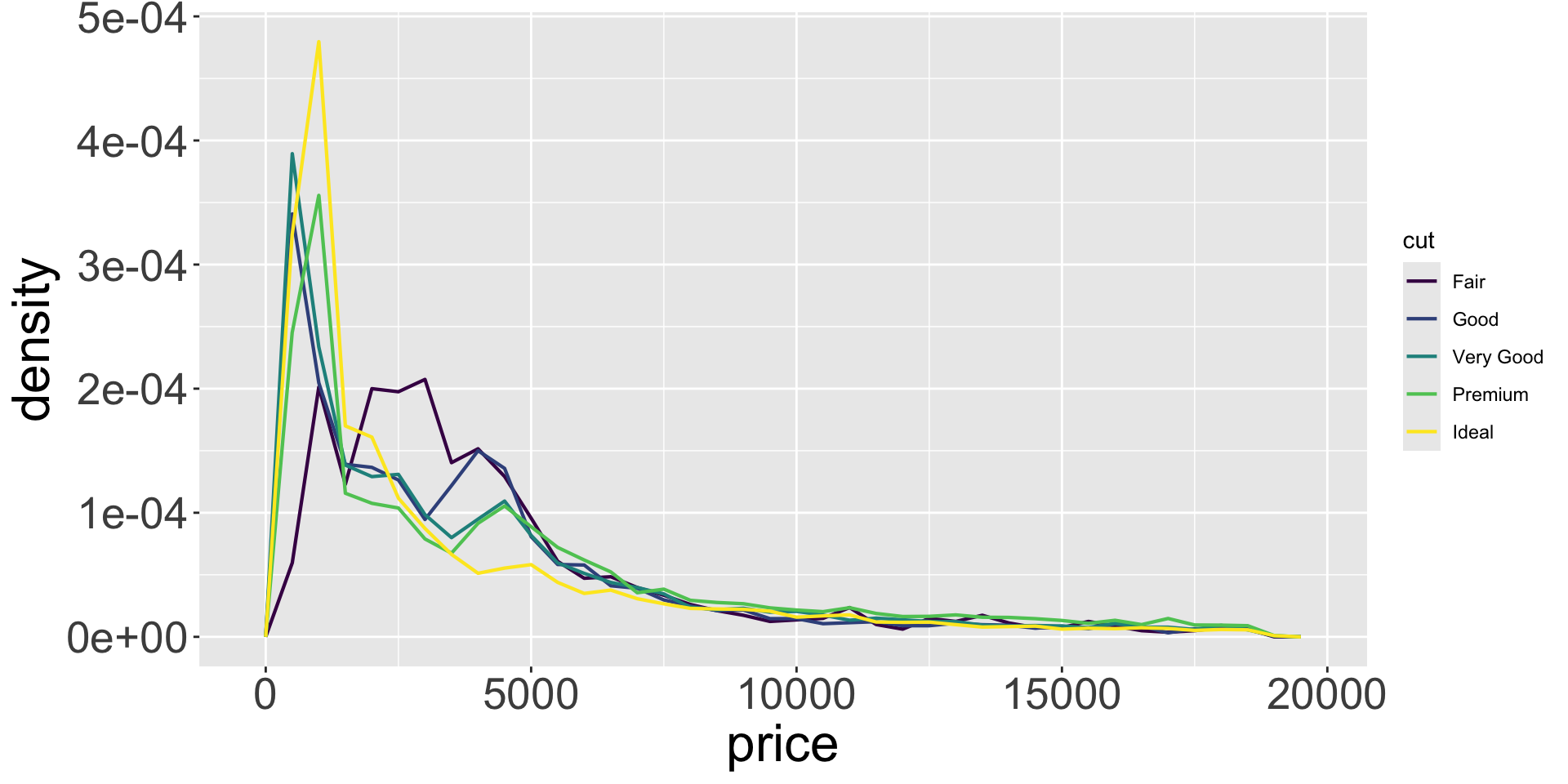
- Seems diamond quality has no significant effect?
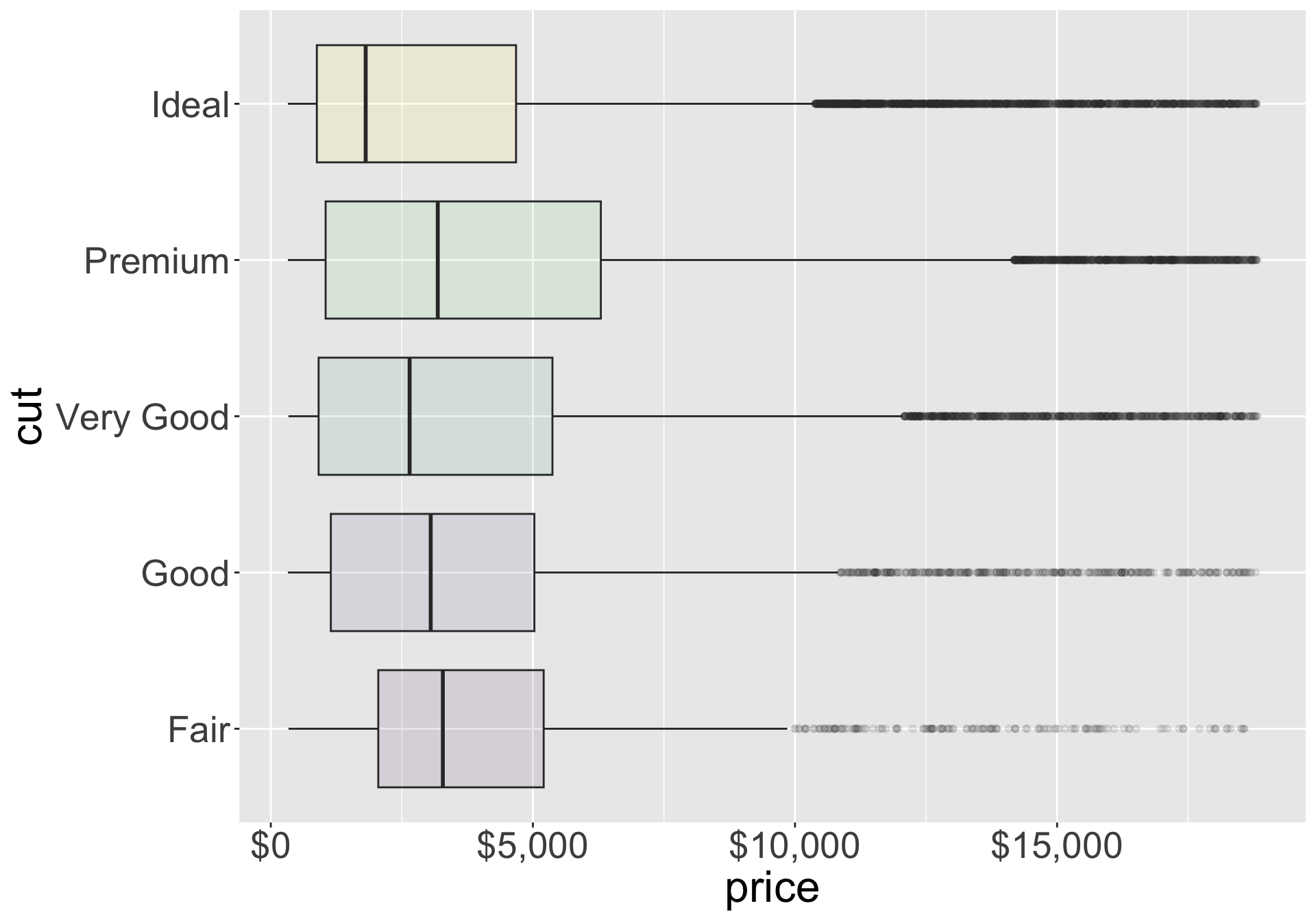
Covariation
- Let’s further inspect with a box plot
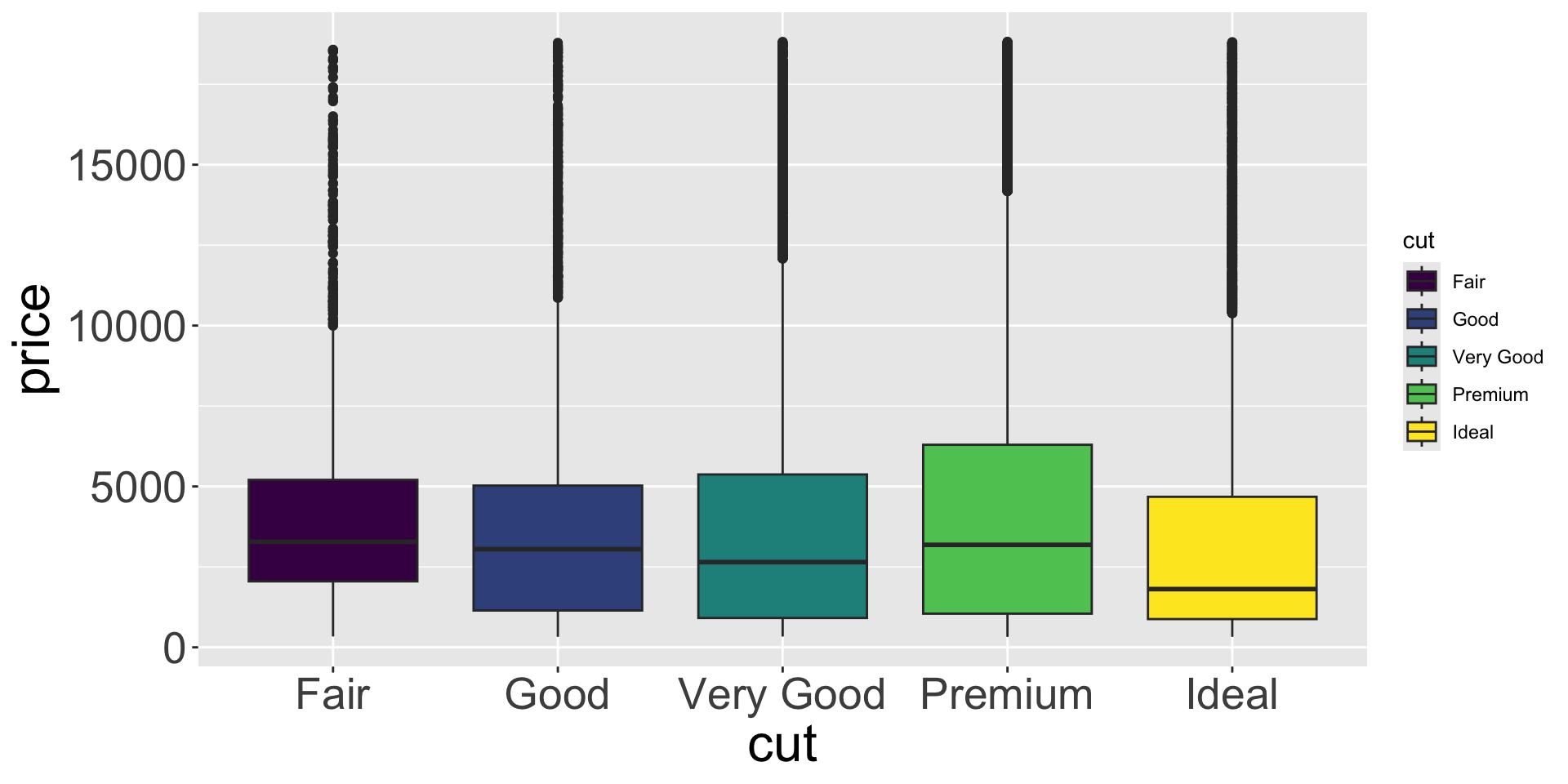
- Can now easily compare medians and 25th/75th percentiles
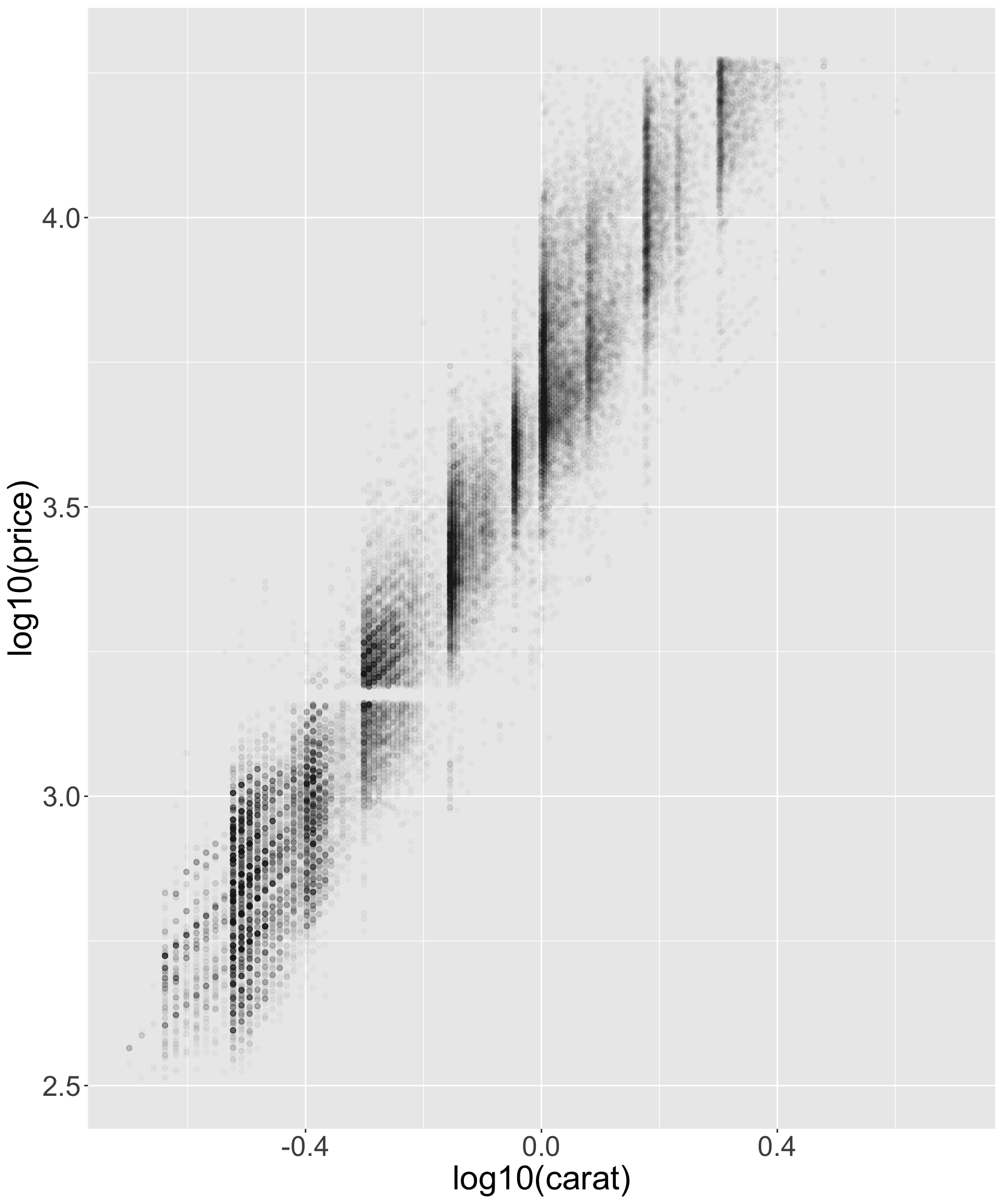
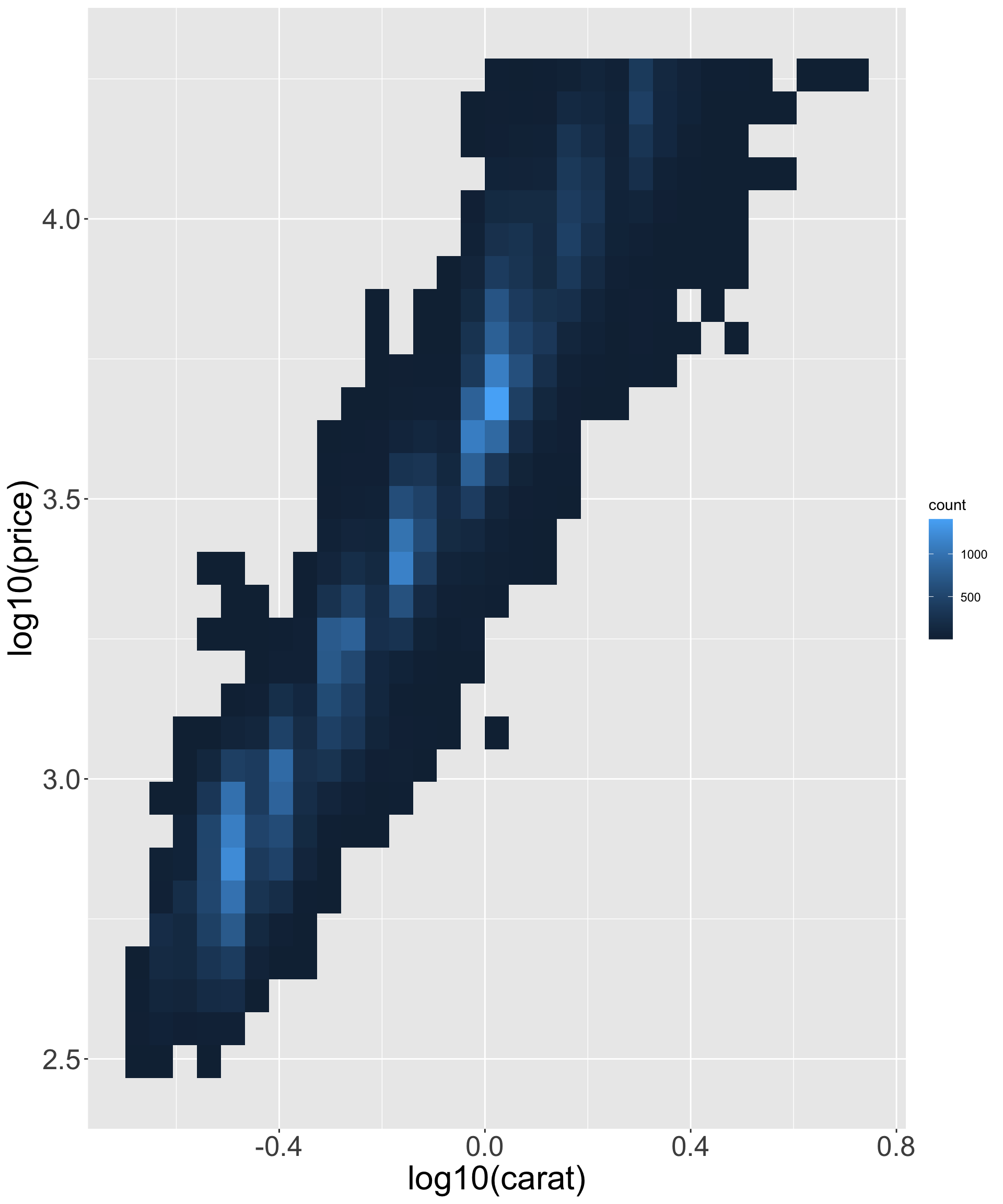
- Better quality diamonds are typically cheaper?! We’ll investigate why.
Better quality diamonds are typically cheaper?!
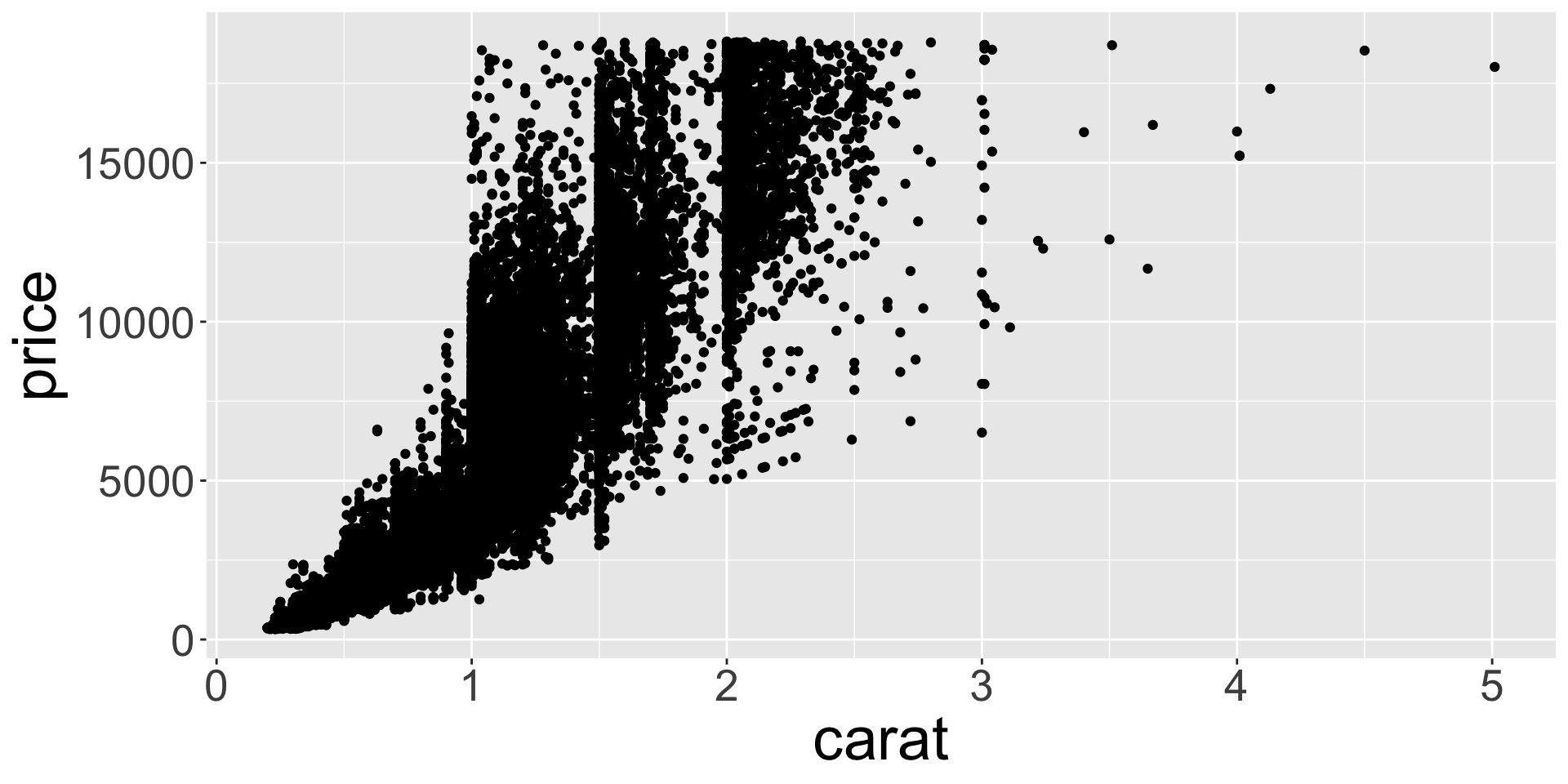
- What might be responsible for this? Recall variables: x,y,z refer to length/width/depth of diamond in mm
- Let’s look at size vs price.
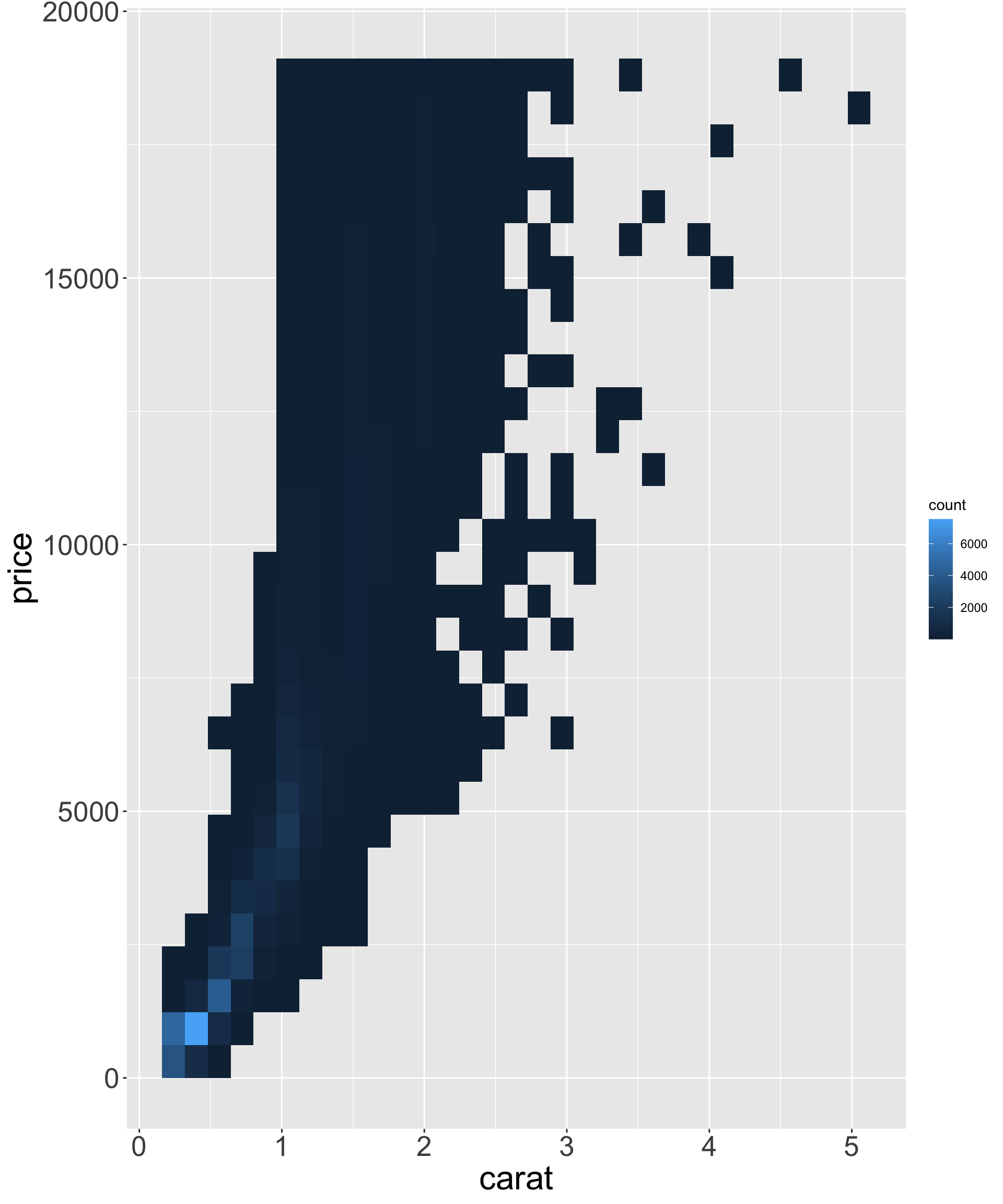
Better quality diamonds are typically cheaper?!
ggplot(diamonds, aes(x, price)) + geom_point() + theme(axis.title = element_text(size=40), axis.text = element_text(size=32))
ggplot(diamonds, aes(y, price)) + geom_point() + theme(axis.title = element_text(size=40), axis.text = element_text(size=32))
ggplot(diamonds, aes(z, price)) + geom_point() + theme(axis.title = element_text(size=40), axis.text = element_text(size=32))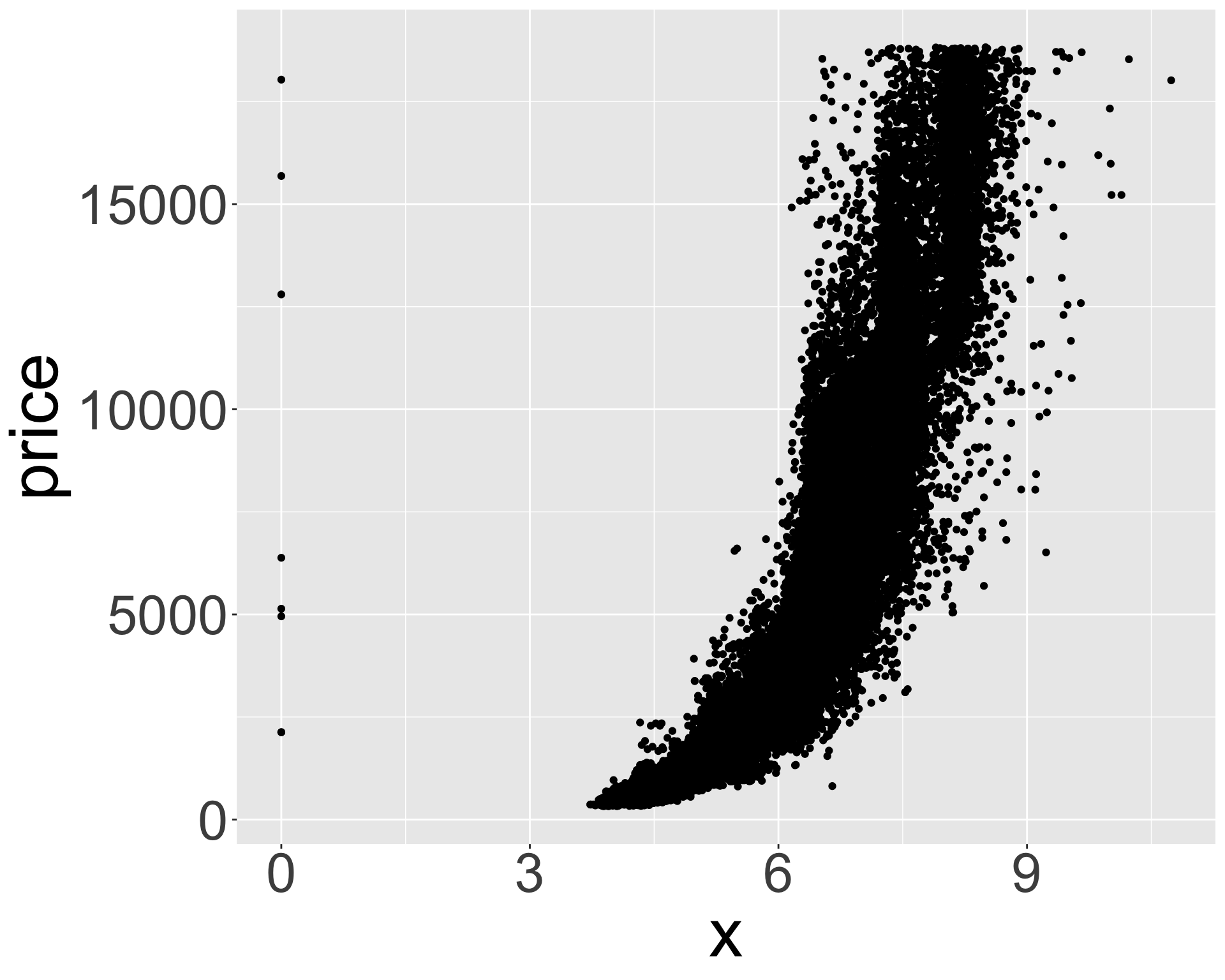
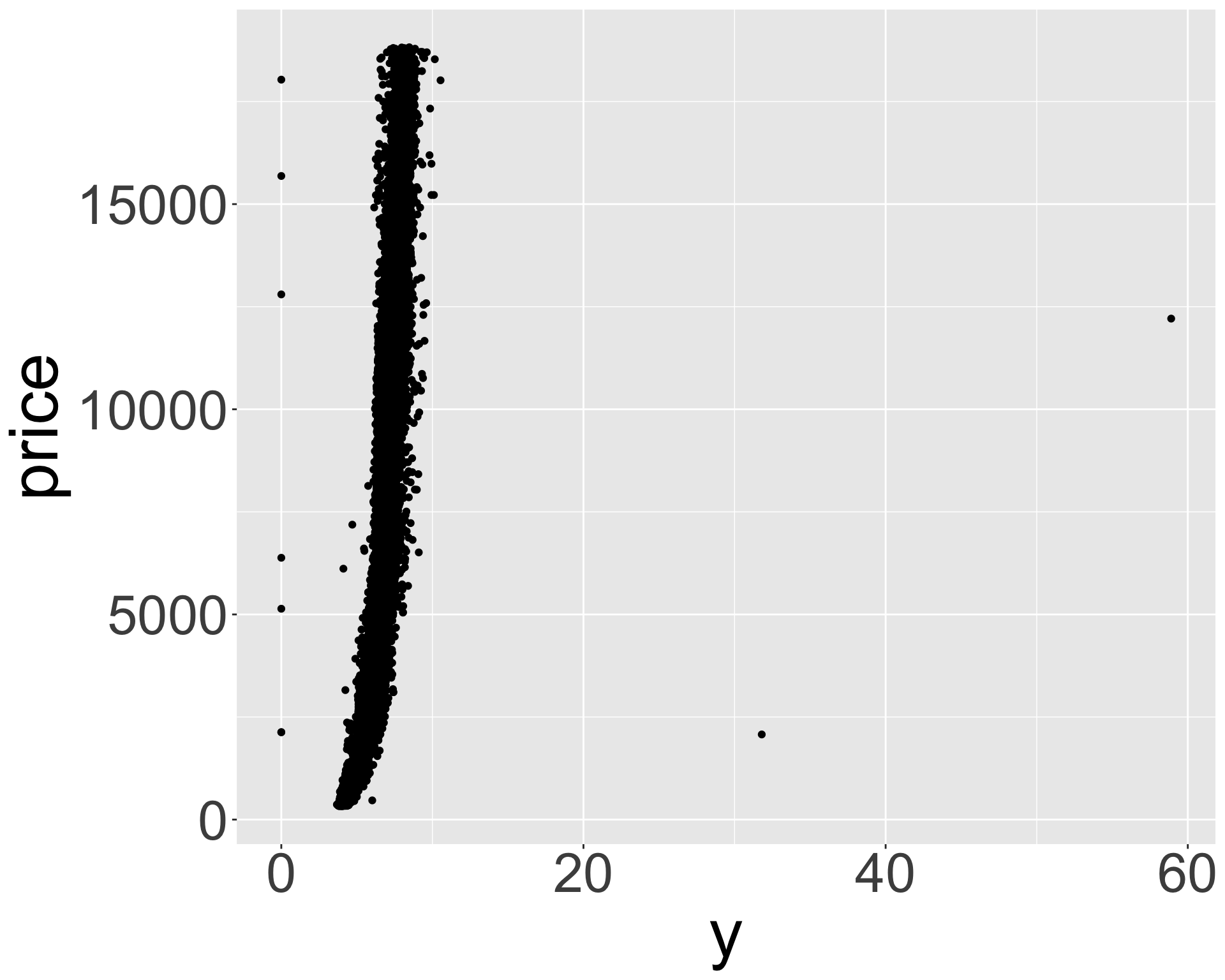
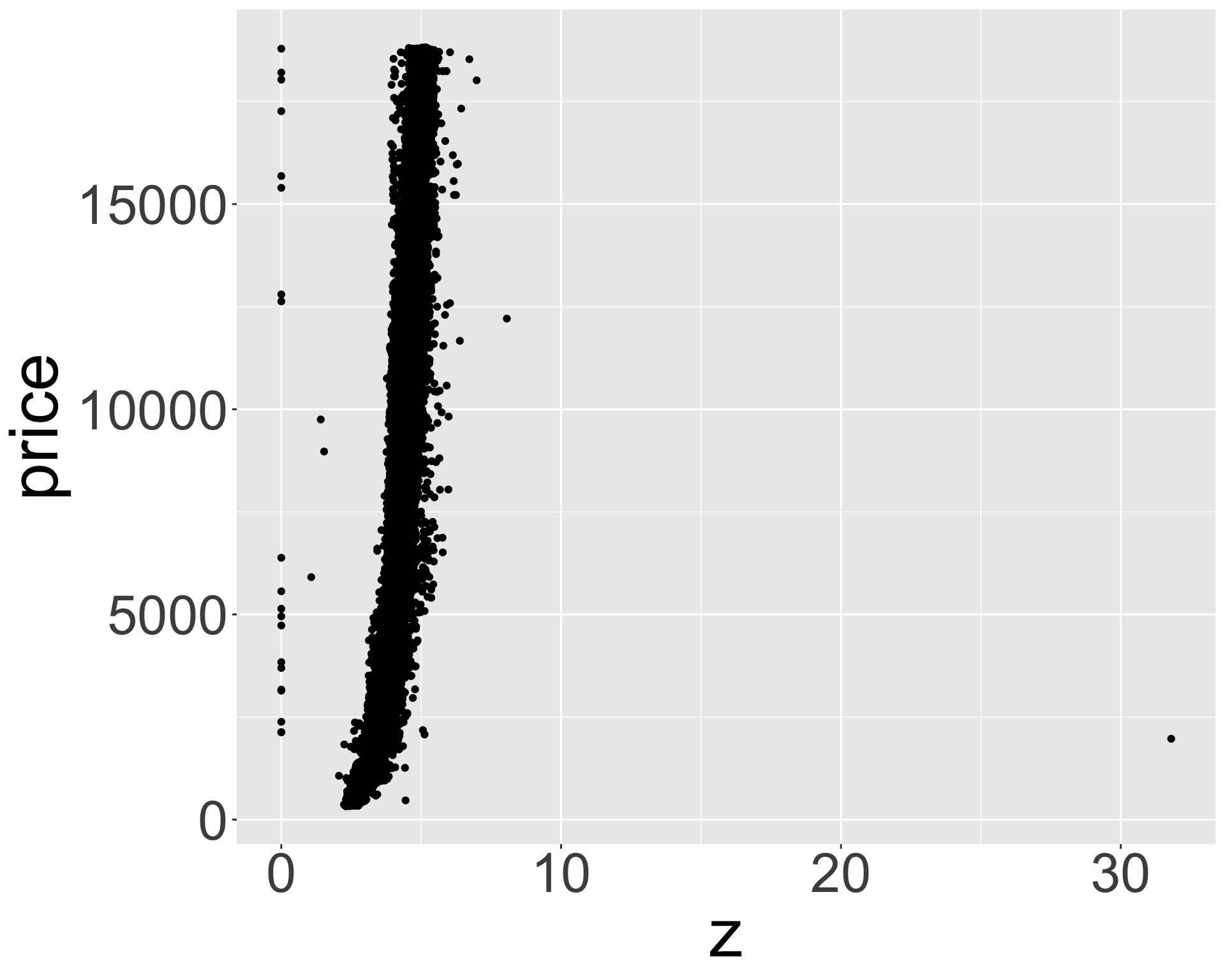
- Clearly some outliers - “zero” length / width / depth diamonds apparently?
- But still some positive correlation between x, y, z and price.
- Let’s clean up the tibble to only view the middle 95% of values for x, y, z.
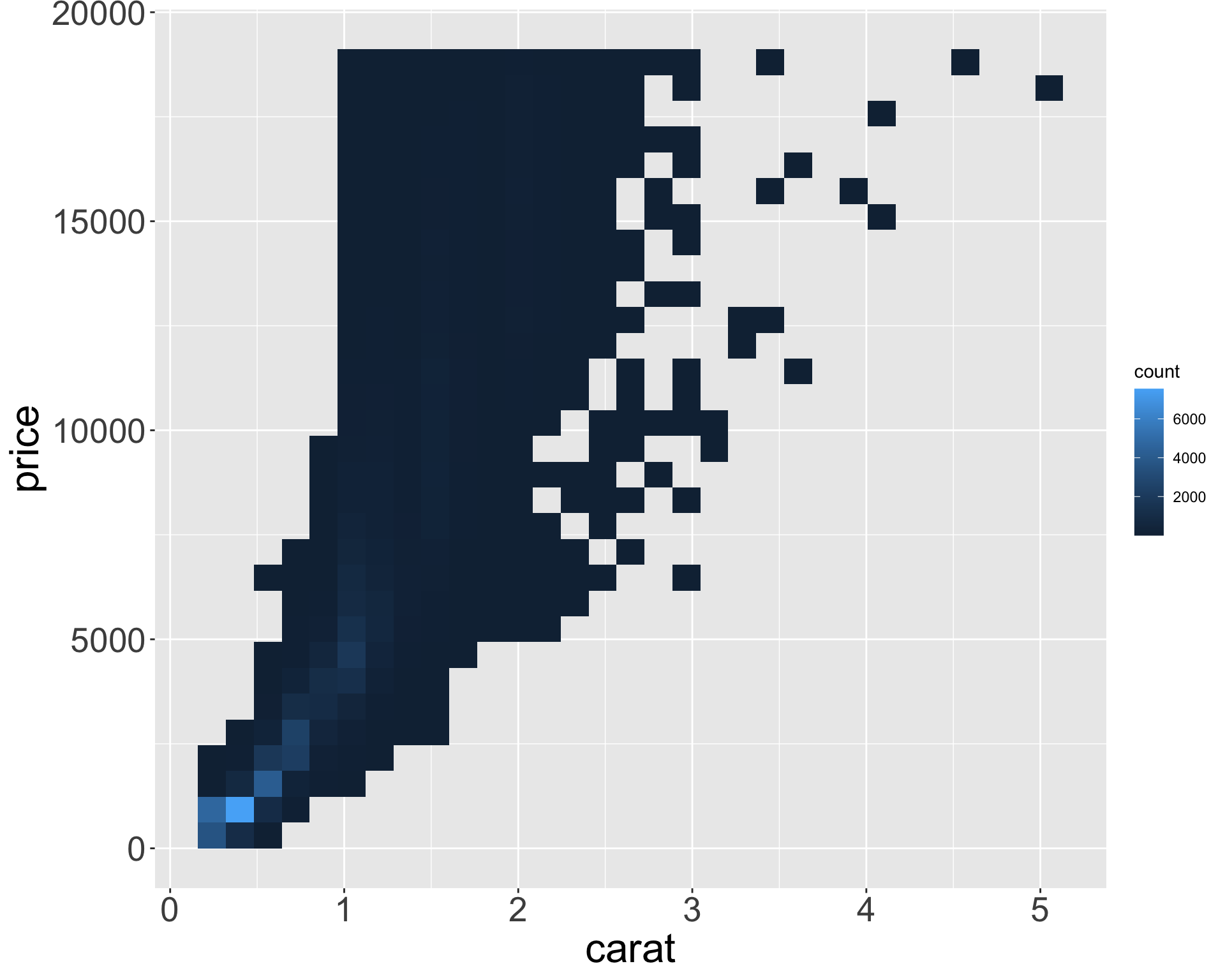
Better quality diamonds are typically cheaper?!
- Let’s clean up the tibble to only view the middle 95% of values for x, y, z
or
Are the two approaches the same?
Better quality diamonds are typically cheaper?!
- Let’s clean up the tibble to only view the middle 95% of values for x, y, z
ggplot(diamonds_middle, aes(x, price)) + geom_point() + theme(axis.title = element_text(size=40), axis.text = element_text(size=32))
ggplot(diamonds_middle, aes(y, price)) + geom_point() + theme(axis.title = element_text(size=40), axis.text = element_text(size=32))
ggplot(diamonds_middle, aes(z, price)) + geom_point() + theme(axis.title = element_text(size=40), axis.text = element_text(size=32))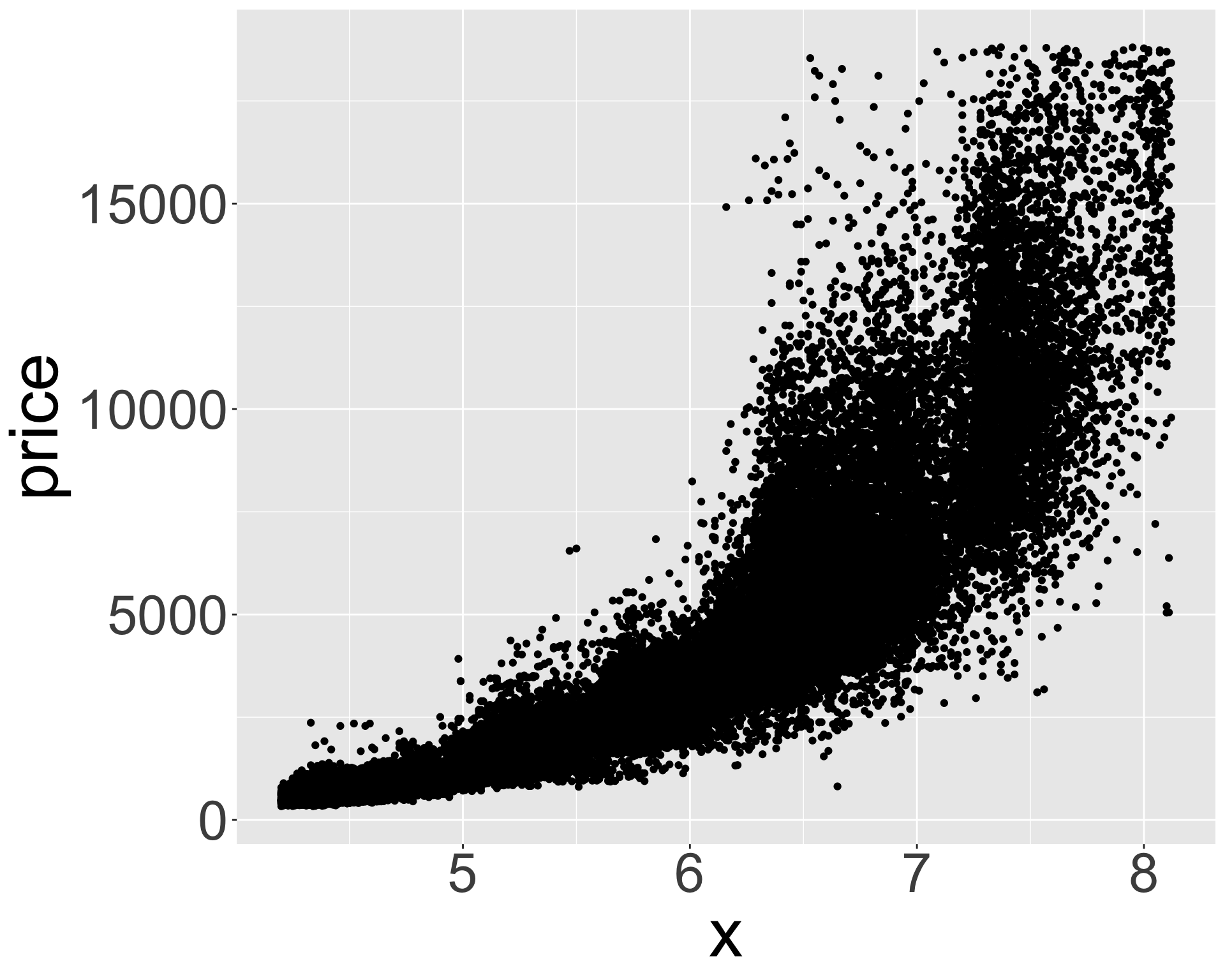
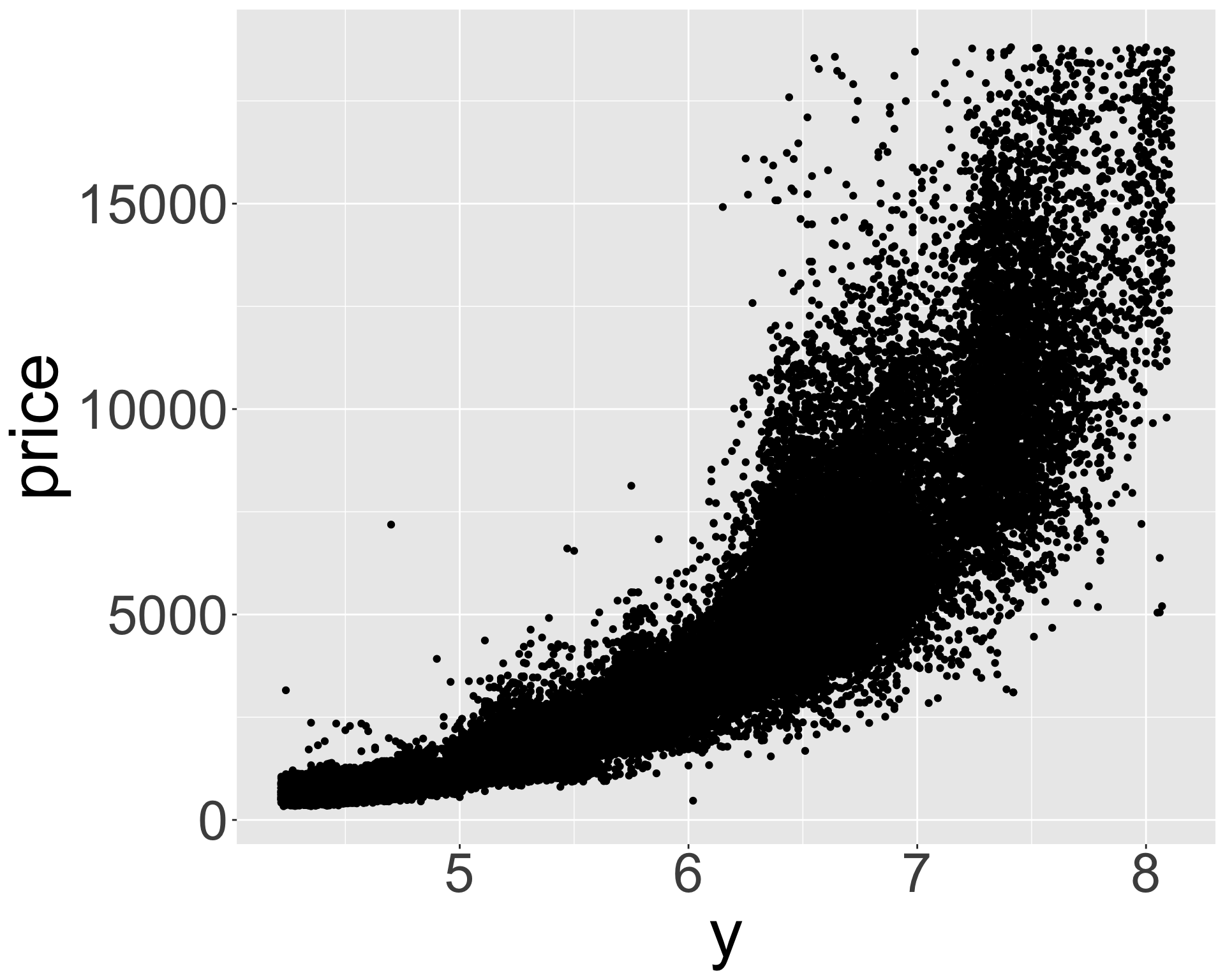
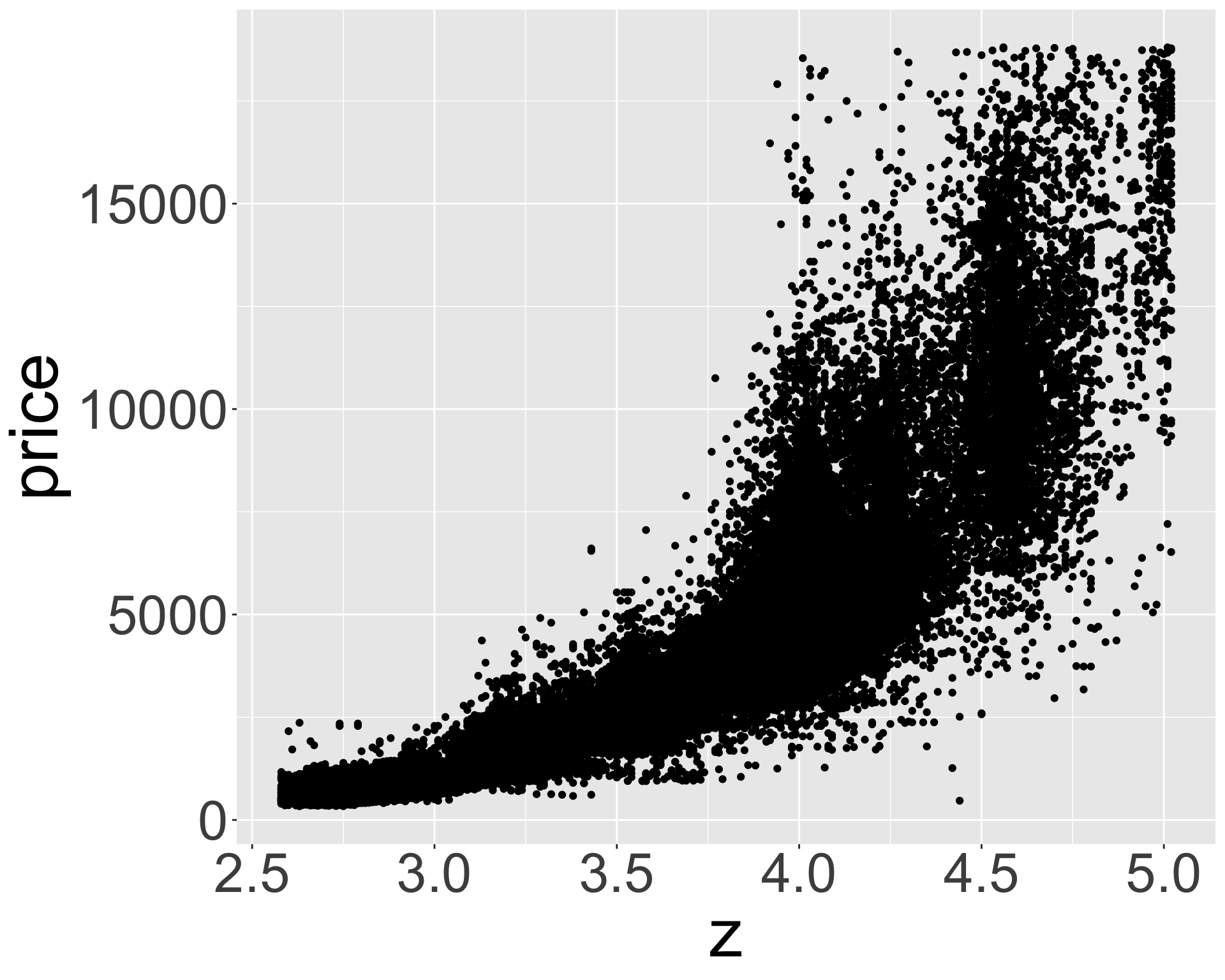
- Appears x, y, z (length/width/depth) are highly correlated with price
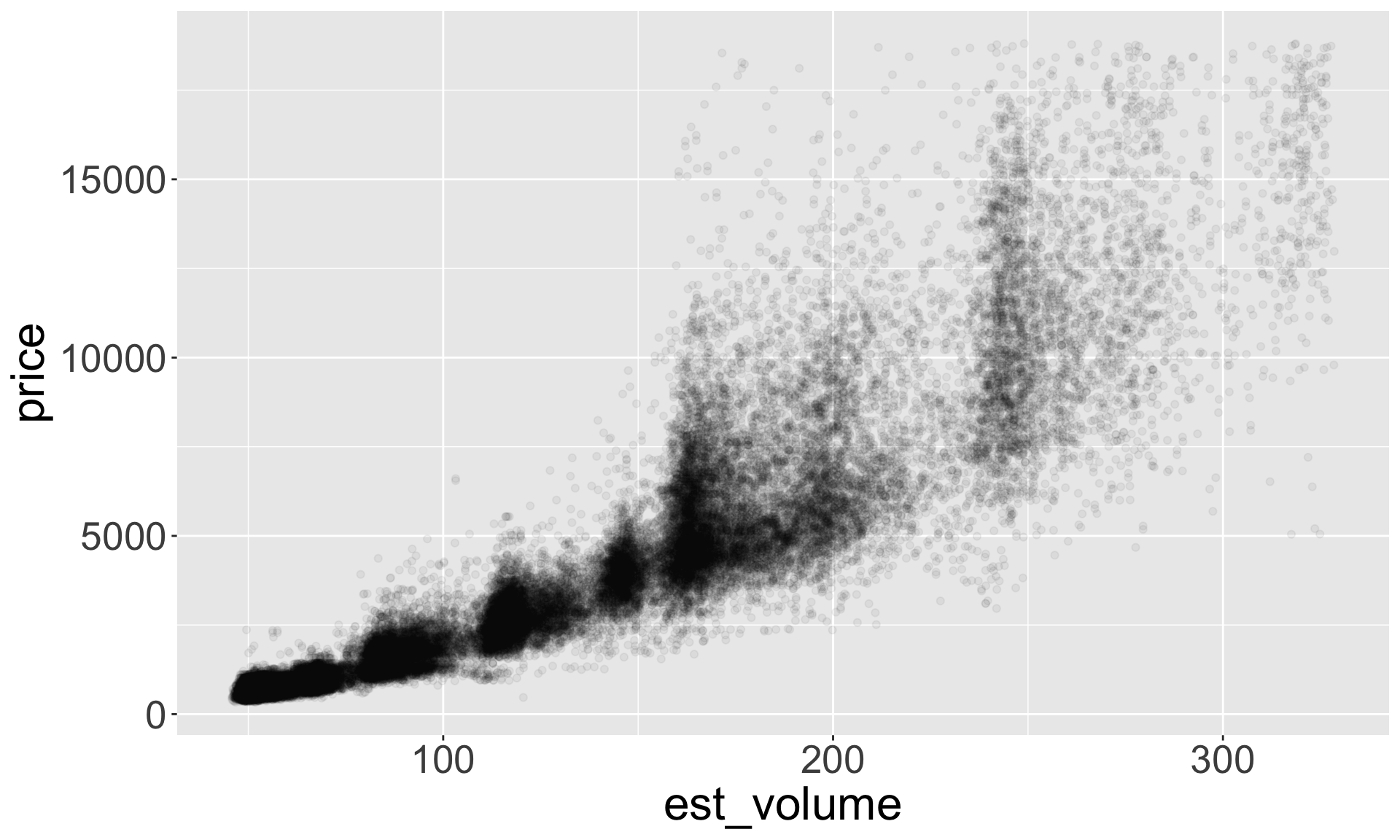
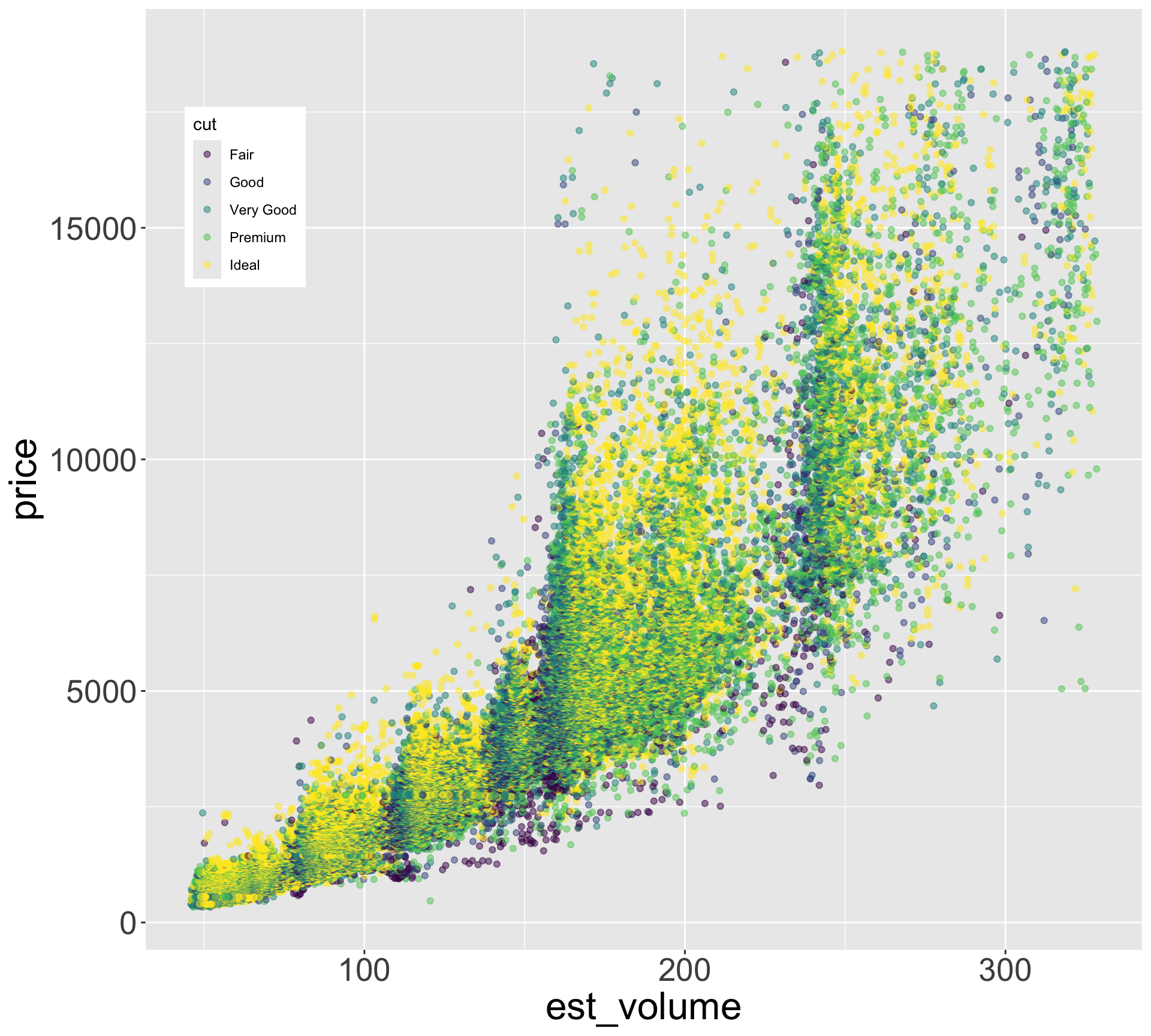
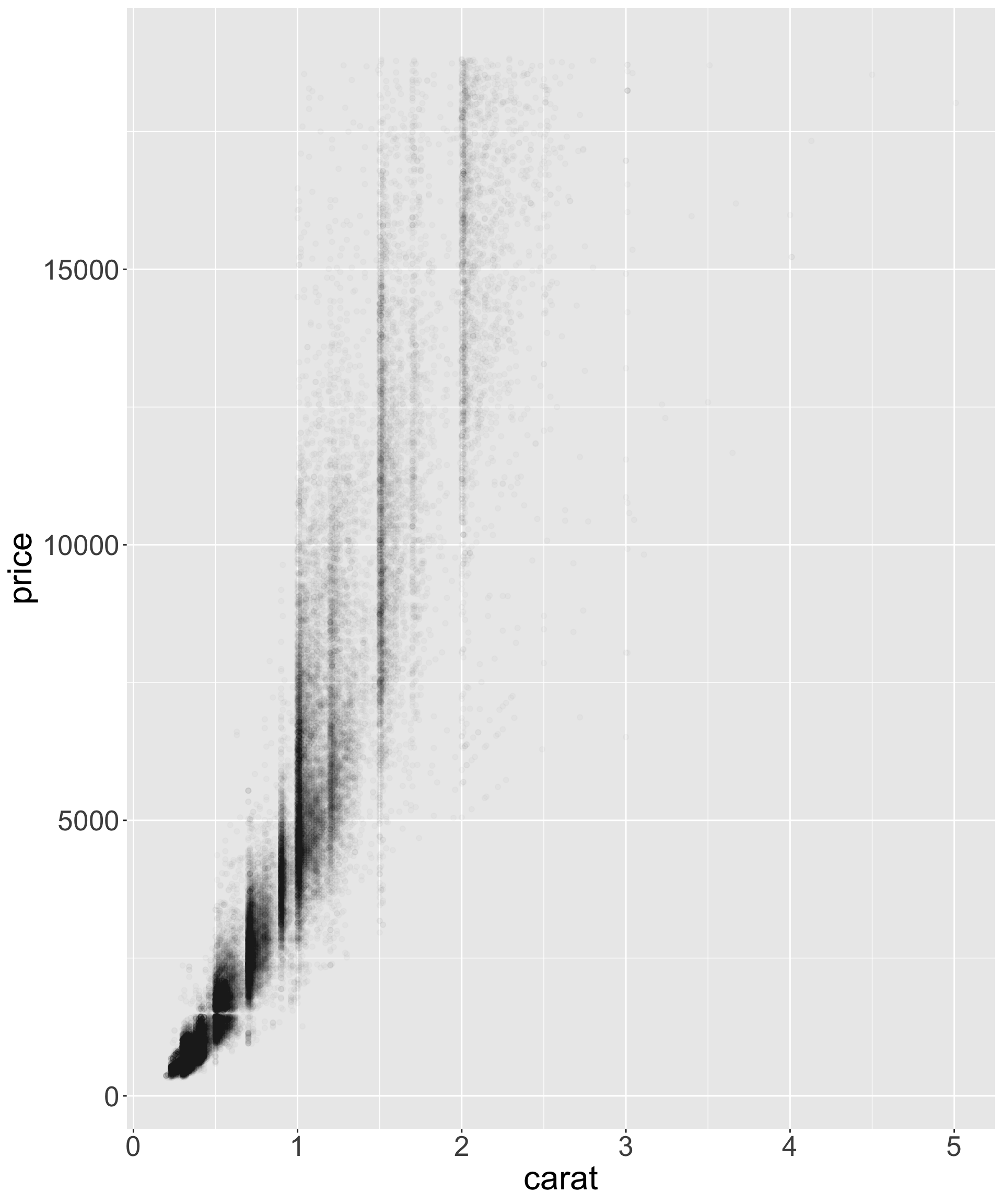
- Let’s explore the relationship between estimated volume (x*y*z) and price
Better quality diamonds are typically cheaper?!
- Let’s look at the relationship between estimated volume (x*y*z) and price
Better quality diamonds are typically cheaper?!
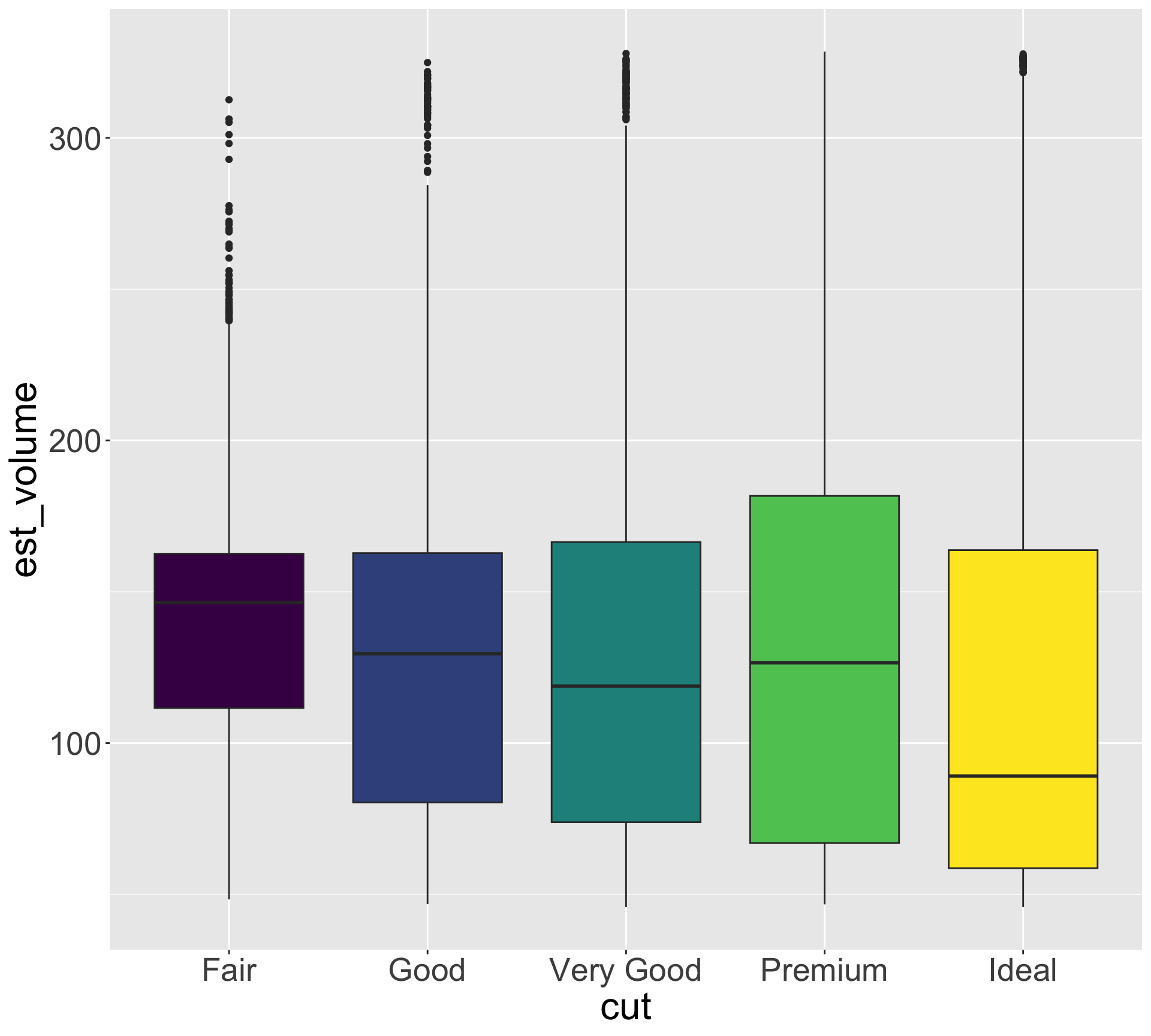
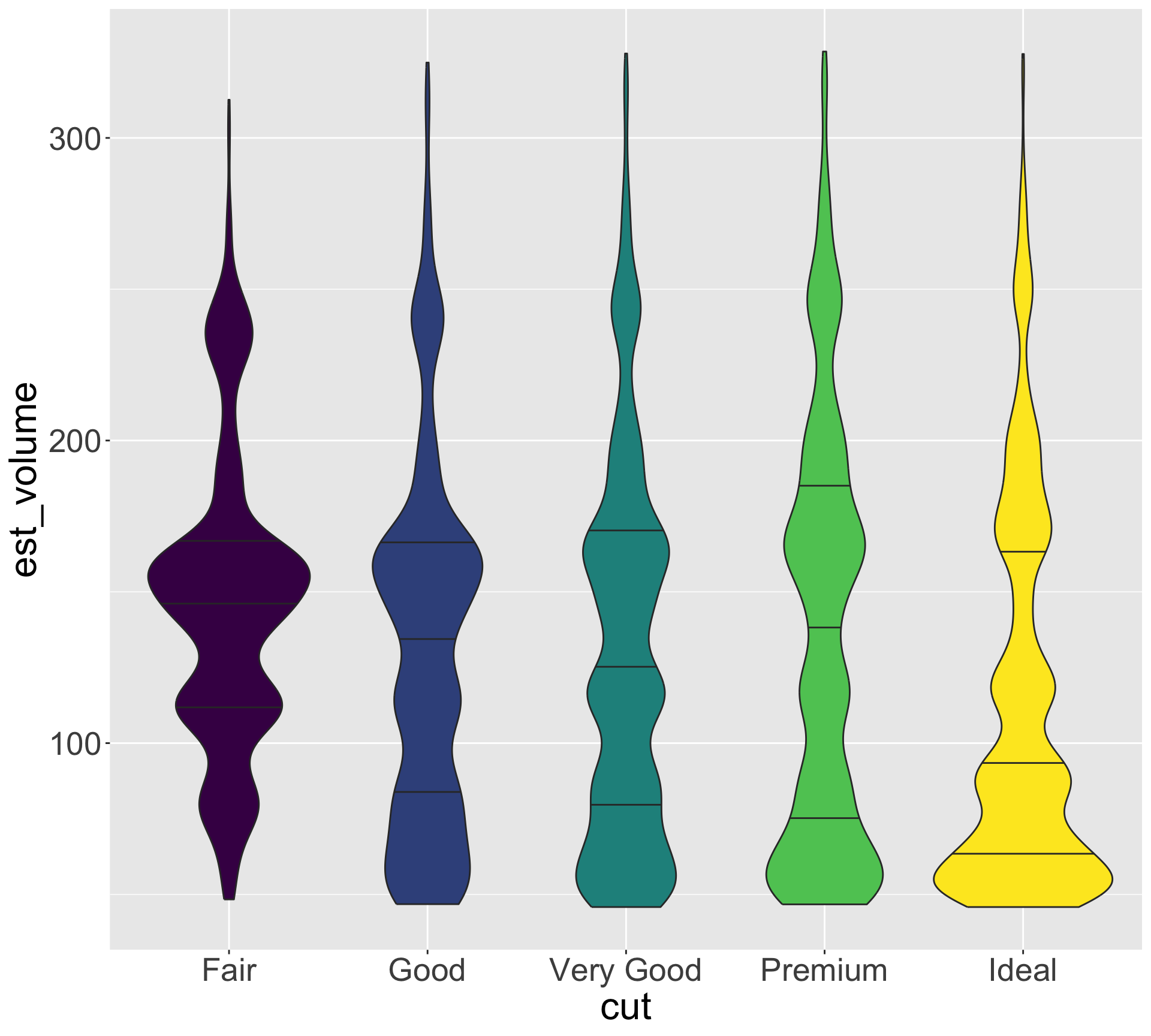
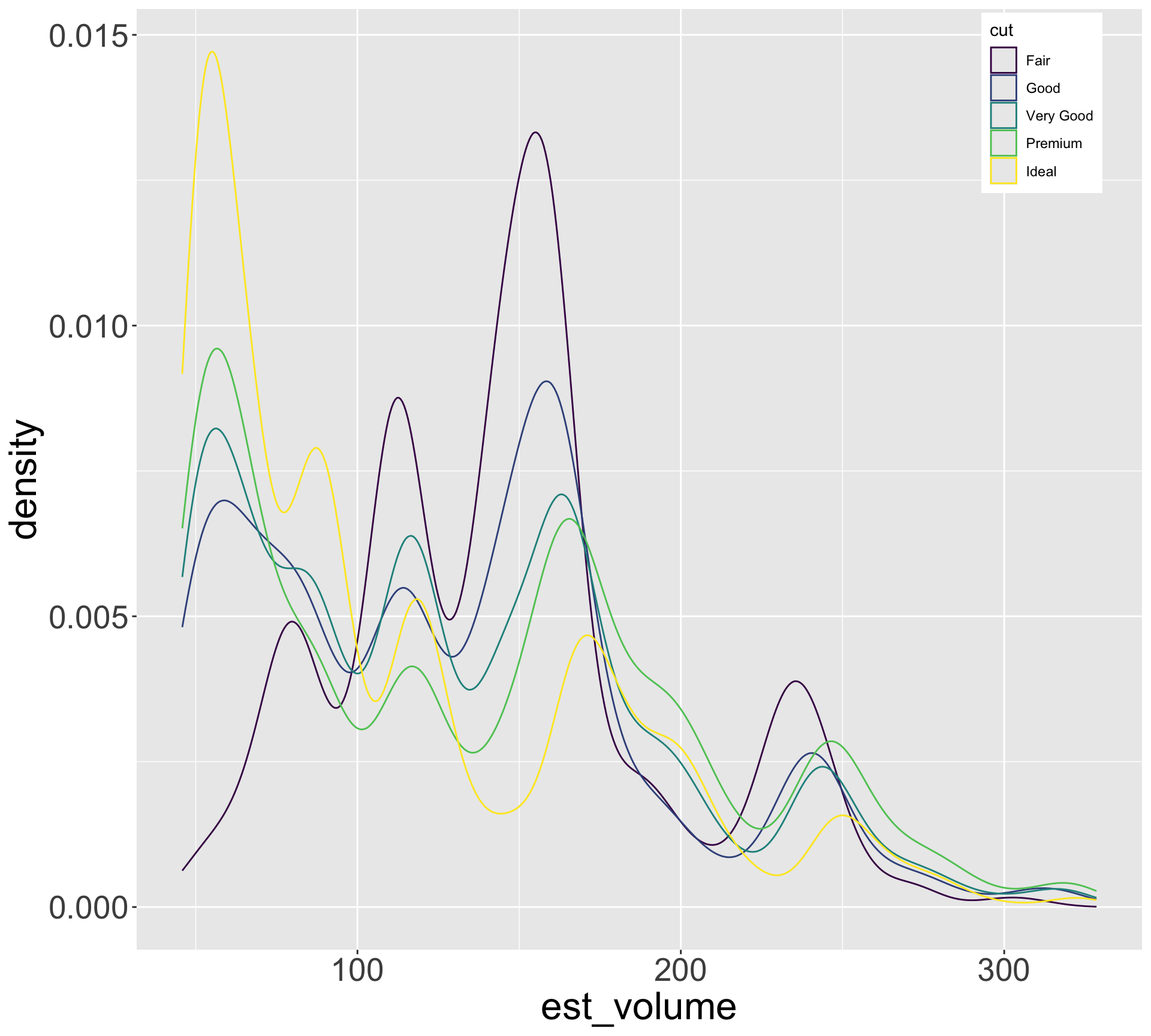
Hypothesis: high quality diamonds are smaller, low quality diamonds are larger.
- To investigate, let’s visualize variation in size for each category of quality
Hypothesis seems reasonable
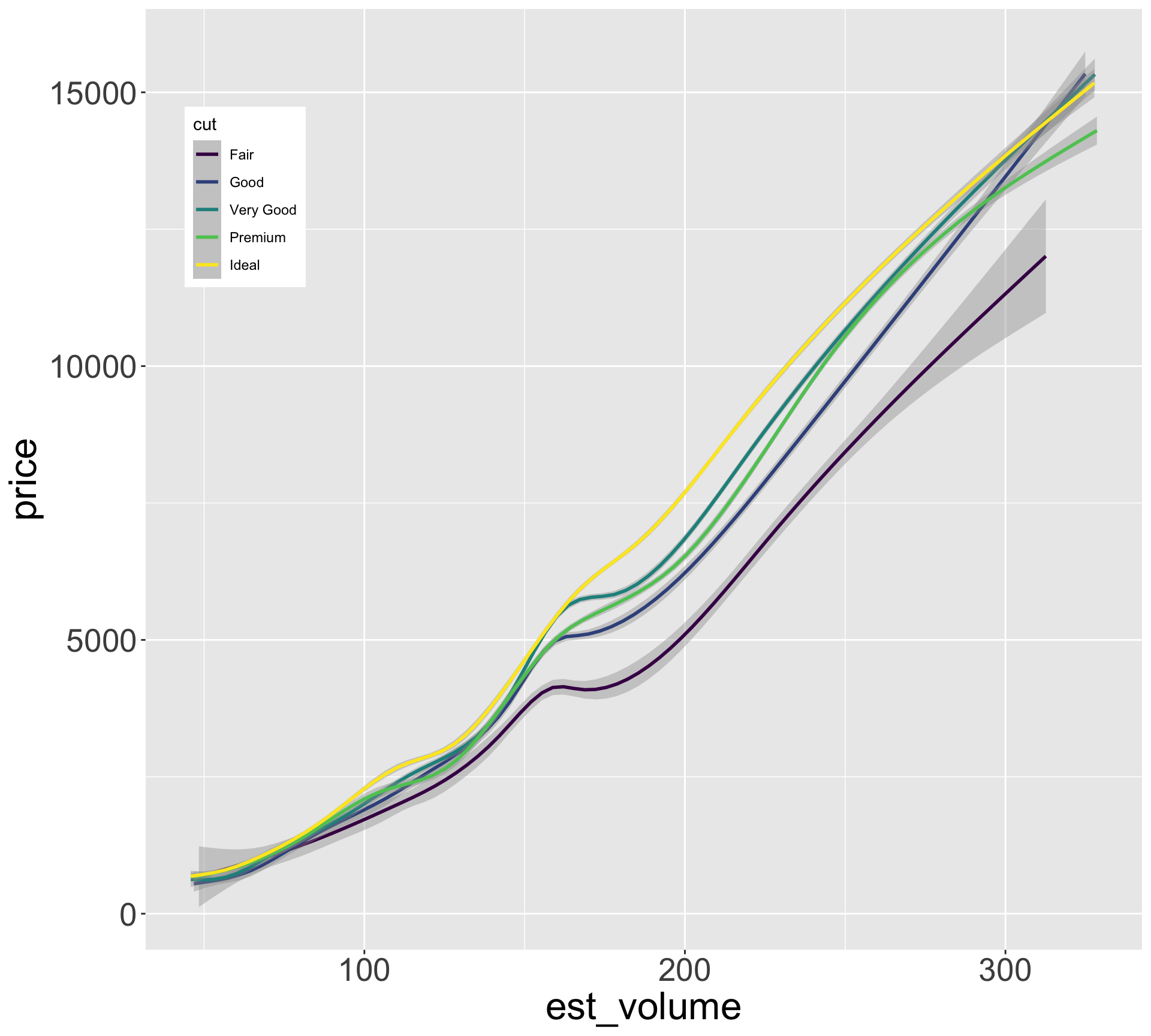
Better quality diamonds are typically cheaper?!
Now let’s look at size vs price for each cut
Communicating ideas
Communicating ideas
- Now we’ll look at how to best display data once we’ve figured out some meaningful relationships between things
- Packages we’ll be using:
tidyverse,scales,ggrepel,patchwork
Communicating ideas
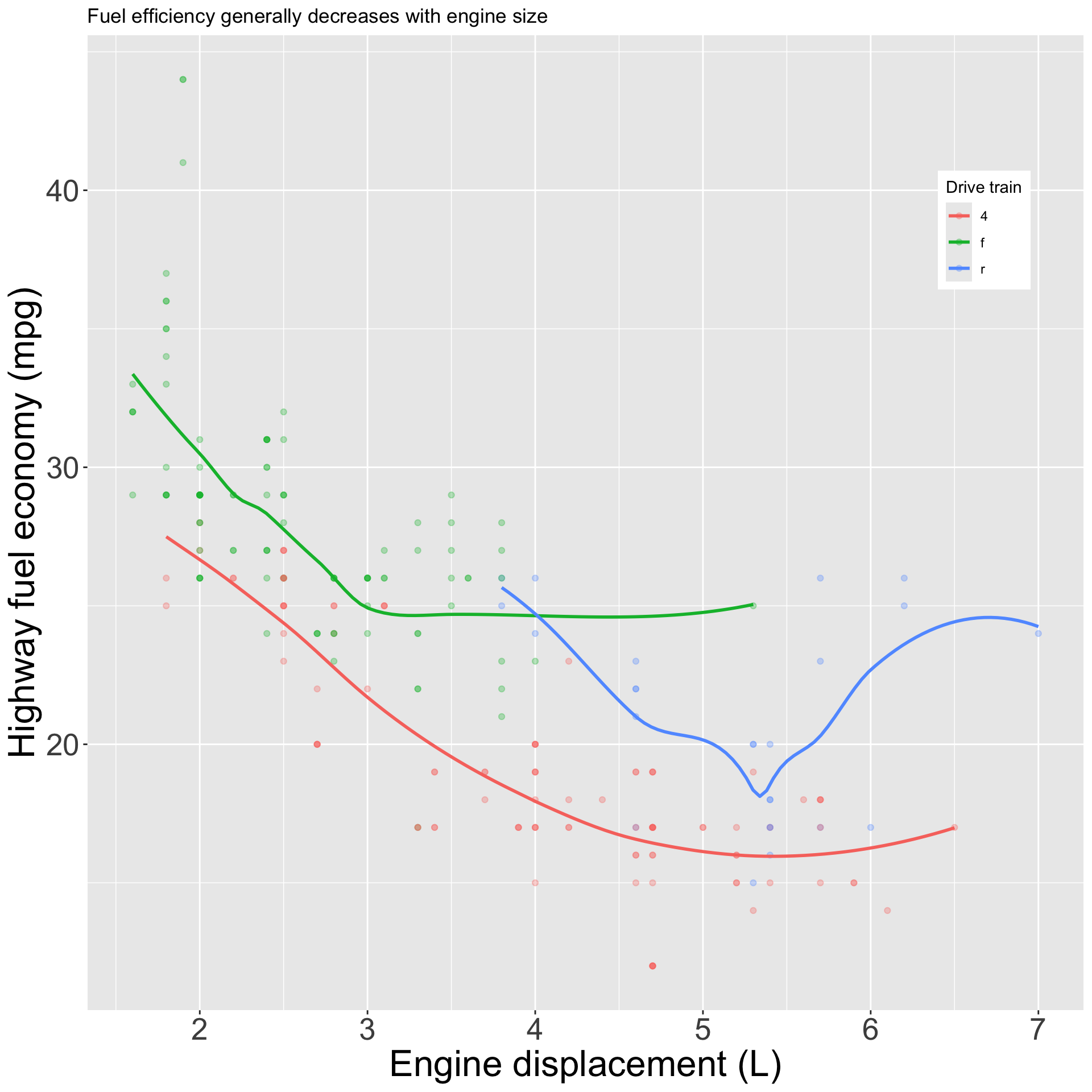
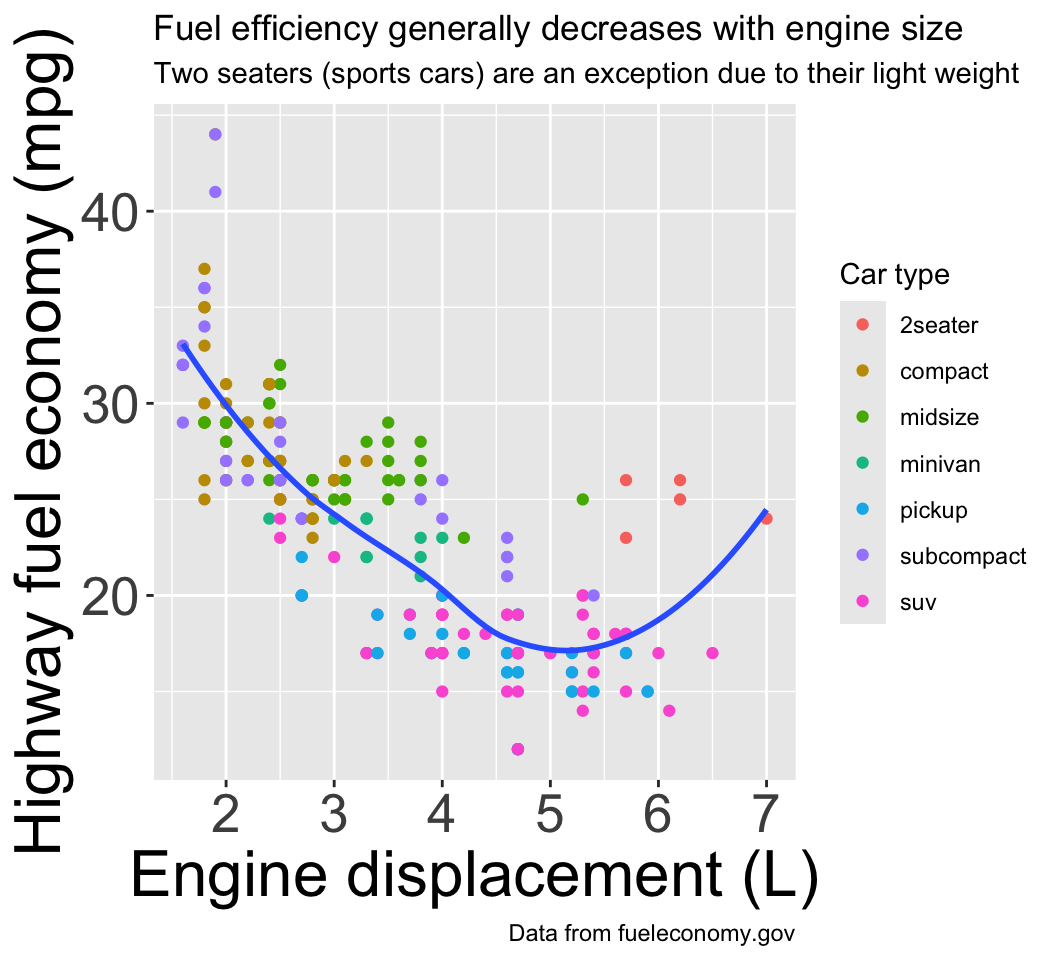
Good labels are crucial to good figures:
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = class)) +
geom_smooth(se = FALSE) +
labs(
x = "Engine displacement (L)",
y = "Highway fuel economy (mpg)",
color = "Car type",
title = "Fuel efficiency generally decreases with engine size",
subtitle = "Two seaters (sports cars) are an exception due to their light weight",
caption = "Data from fueleconomy.gov"
)
Annotations
Annotations
Often helpful to label individual observations or groups of observations
geom_text(), similar togeom_point()but has additional aestheticlabel.- Label could come from tibble itself, or could use custom user-added labels
- Let’s create custom labels
Annotations
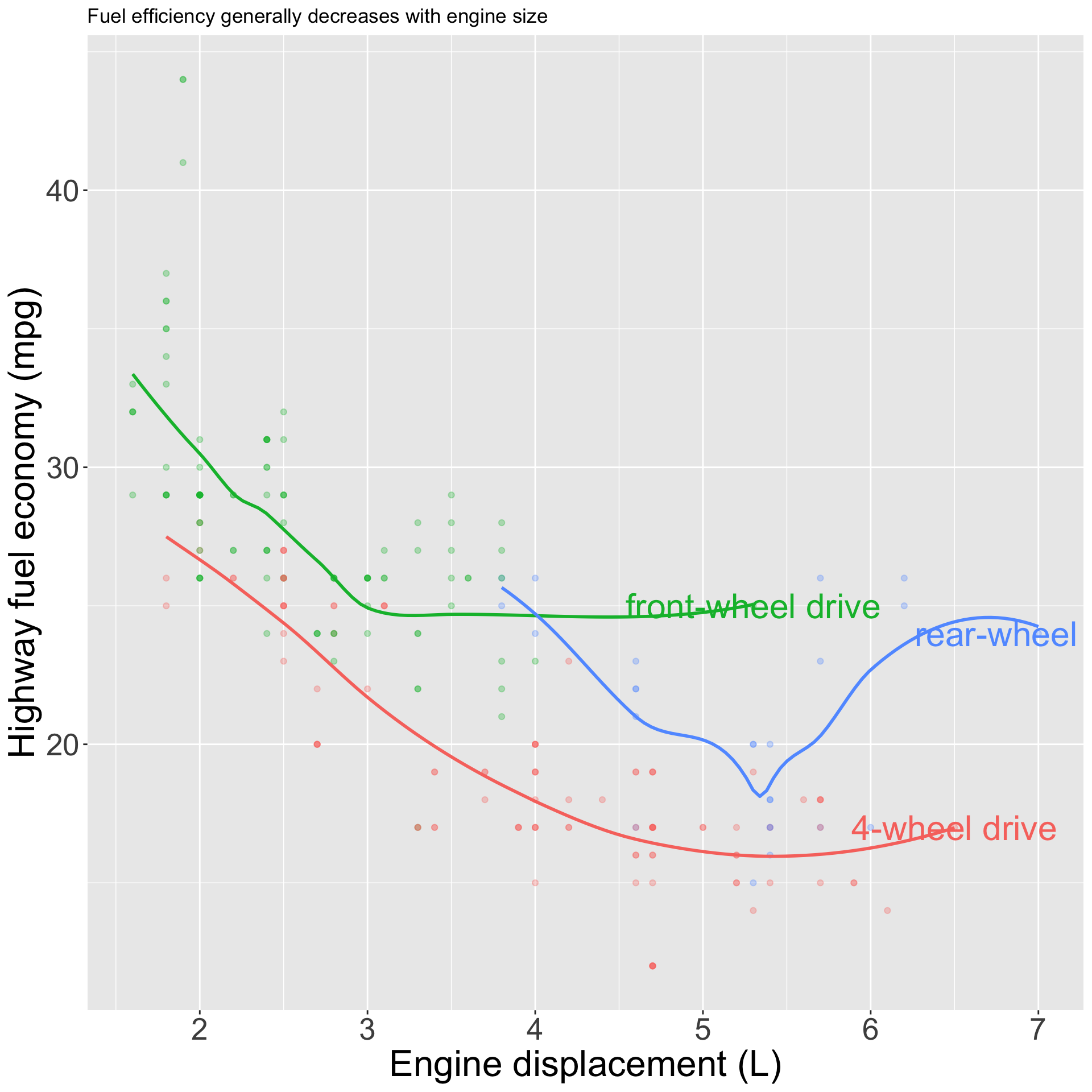
pp <- mpg |>
ggplot(aes(displ, hwy, color = drv)) +
geom_point(alpha = 0.3) +
geom_smooth(se = FALSE) +
geom_text(
data = label_info,
aes(label = drive_type),
size = 8
) +
theme(legend.position = "none") +
labs(x = "Engine displacement (L)", y = "Highway fuel economy (mpg)", color = "Drive train", title = "Fuel efficiency generally decreases with engine size")Annotations
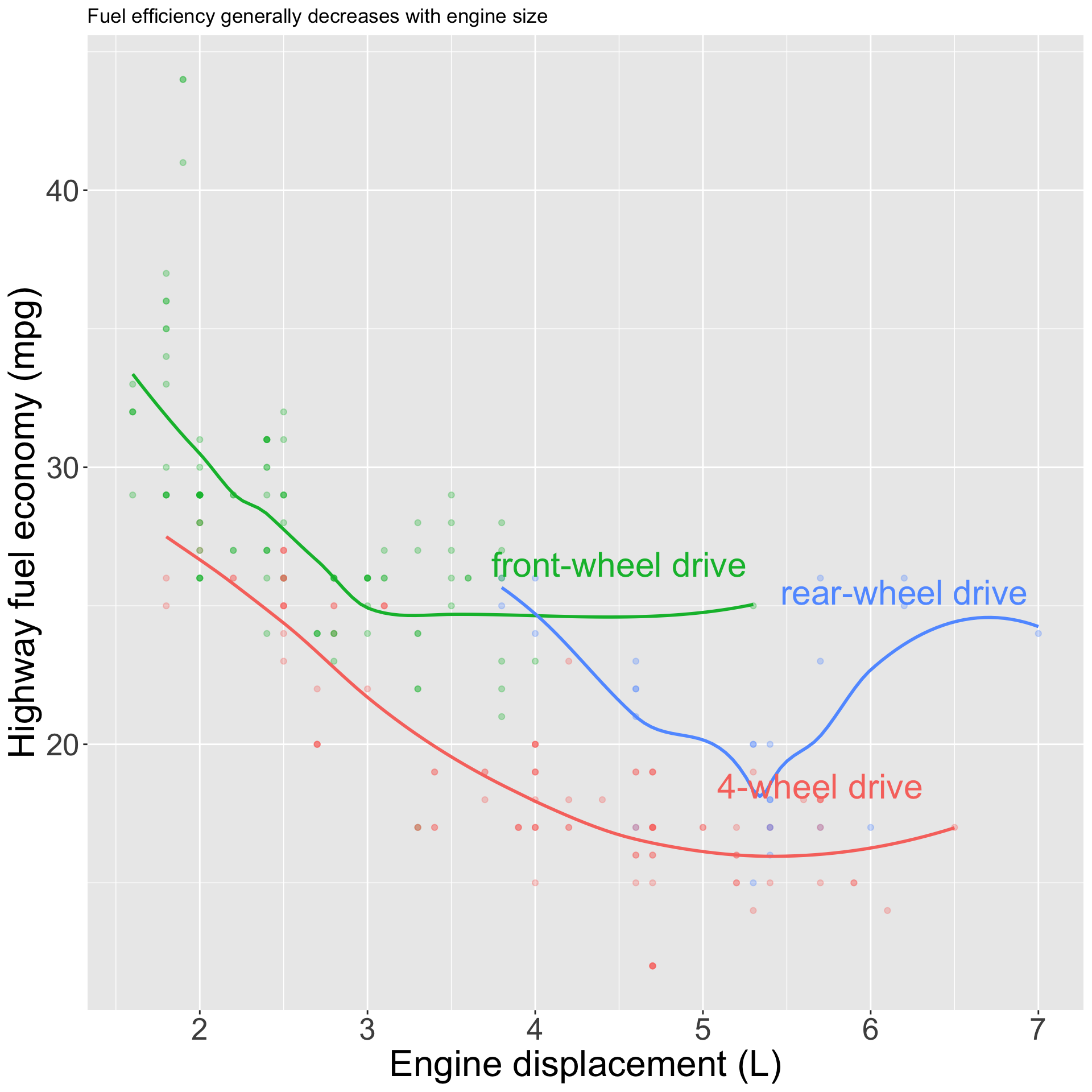
pp <- mpg |>
ggplot(aes(displ, hwy, color = drv)) +
geom_point(alpha = 0.3) +
geom_smooth(se = FALSE) +
geom_text(
data = label_info,
aes(label = drive_type),
nudge_y = 1.5, nudge_x = -0.8,
size = 8
) +
theme(legend.position = "none") +
labs(x = "Engine displacement (L)", y = "Highway fuel economy (mpg)", color = "Drive train", title = "Fuel efficiency generally decreases with engine size")Annotations
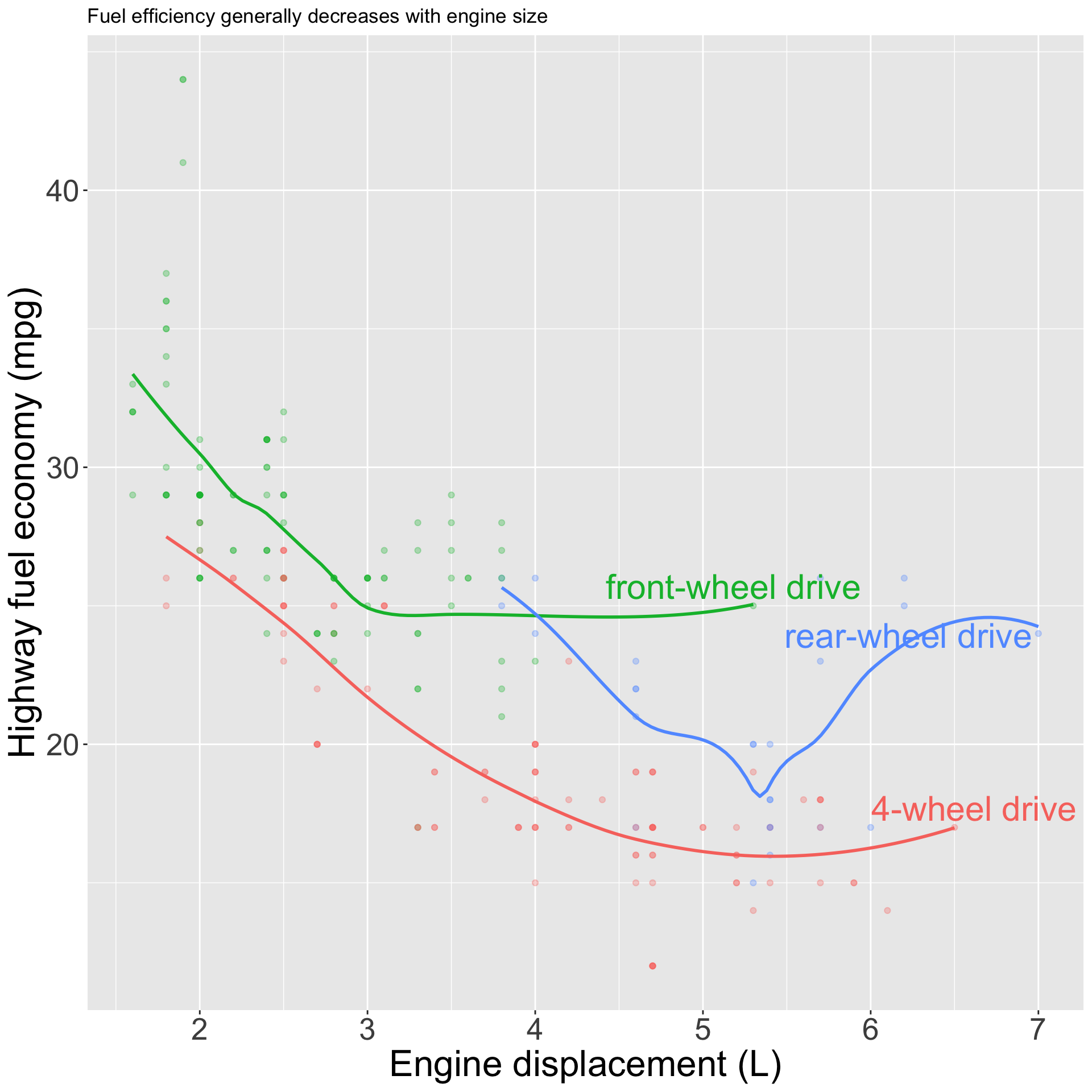
pp <- mpg |>
ggplot(aes(displ, hwy, color = drv)) +
geom_point(alpha = 0.3) +
geom_smooth(se = FALSE) +
geom_text_repel(
data = label_info,
aes(label = drive_type),
size = 8
) +
theme(legend.position = "none") +
labs(x = "Engine displacement (L)", y = "Highway fuel economy (mpg)", color = "Drive train", title = "Fuel efficiency generally decreases with engine size")Annotations
Same idea can be used to highlight points on plot, e.g. outliers
potential_outliers <- mpg |> filter(hwy > 40 | (hwy > 20 & displ > 5))
ggplot(mpg, aes(displ, hwy)) +
geom_point() +
geom_text_repel(data = potential_outliers, aes(label = model)) +
geom_point(data = potential_outliers, shape = "circle open", color = "red", size = 3) +
labs(x = "Engine displacement (L)", y = "Highway fuel economy (mpg)", color = "Drive train", title = "Fuel efficiency generally decreases with engine size")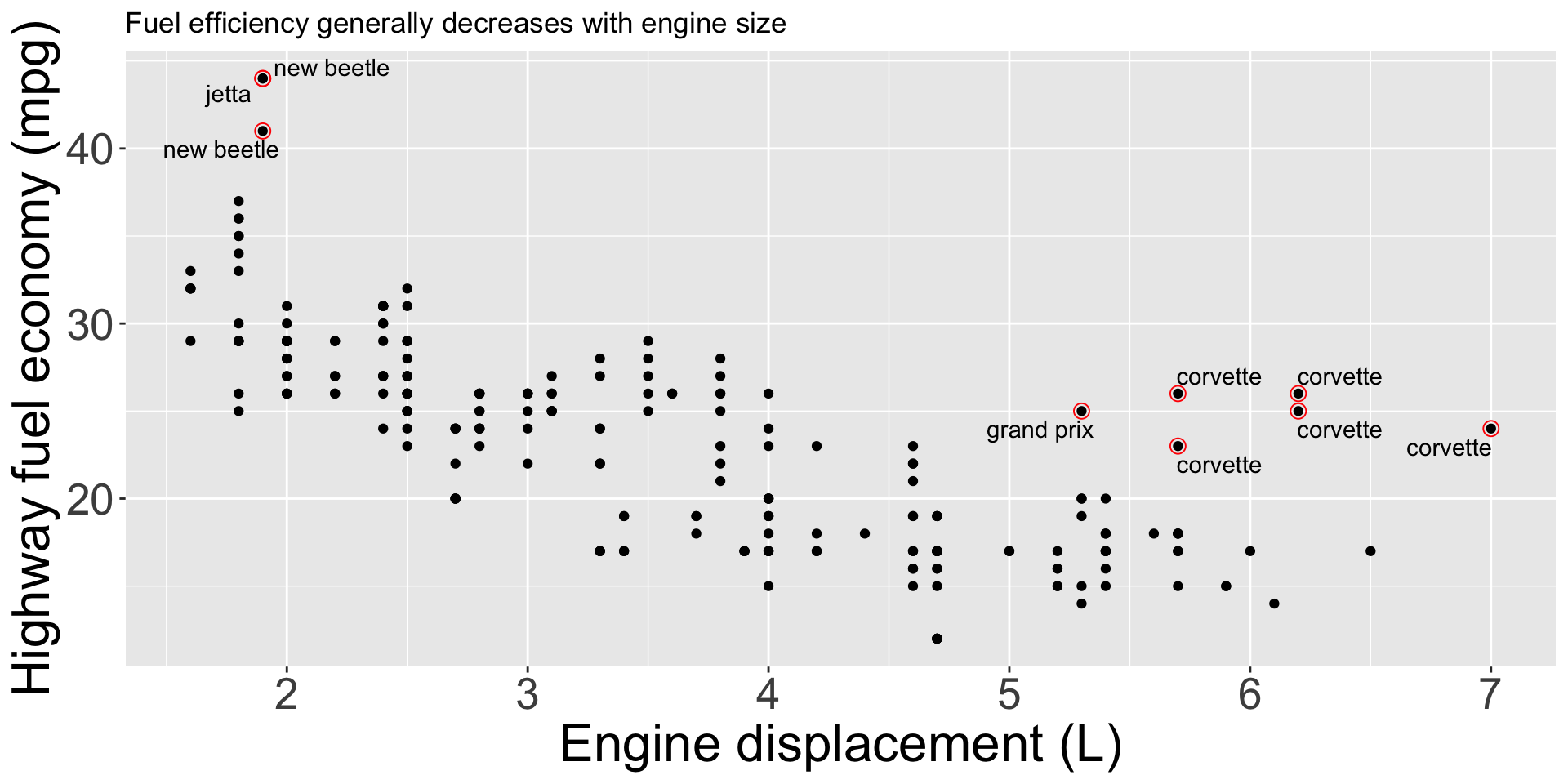
Annotations
- Other annotations which you might encounter / could be useful:
geom_hline(),geom_vline(): create horizontal / vertical linesgeom_rect(): rectangle around points of interest, require specifyingxmin,xmax,ymin,ymaxgeom_segment(): draw an arrow from one point to another
- Function which allows for doing any of these:
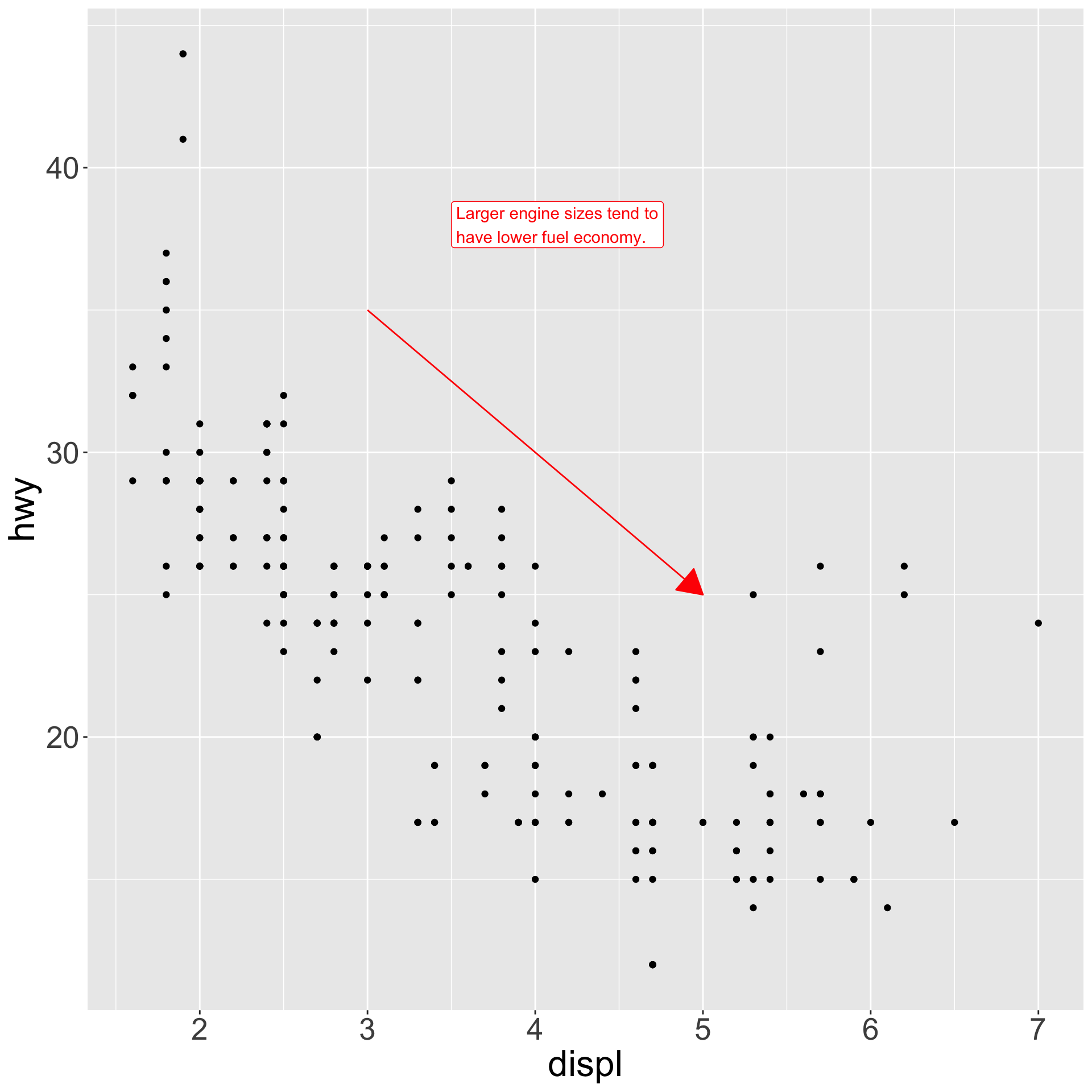
annotate() - Let’s say we want to add some text to our plot, but split lines every 30 characters using
str_wrap()
Annotations
- Let’s add a text label in red using
annotate(geom = 'label') - … and then add a line segment giving general trend
Axis ticks
Axes and legends are called guides in R.
- Axes used for x,y aesthetics, legends for everything else
- Ticks on axes and keys on legend affected by args
breaksandlabels.
Axis ticks
- Specifying
labelsandbreaksprovides a lot of flexibility on how to plot
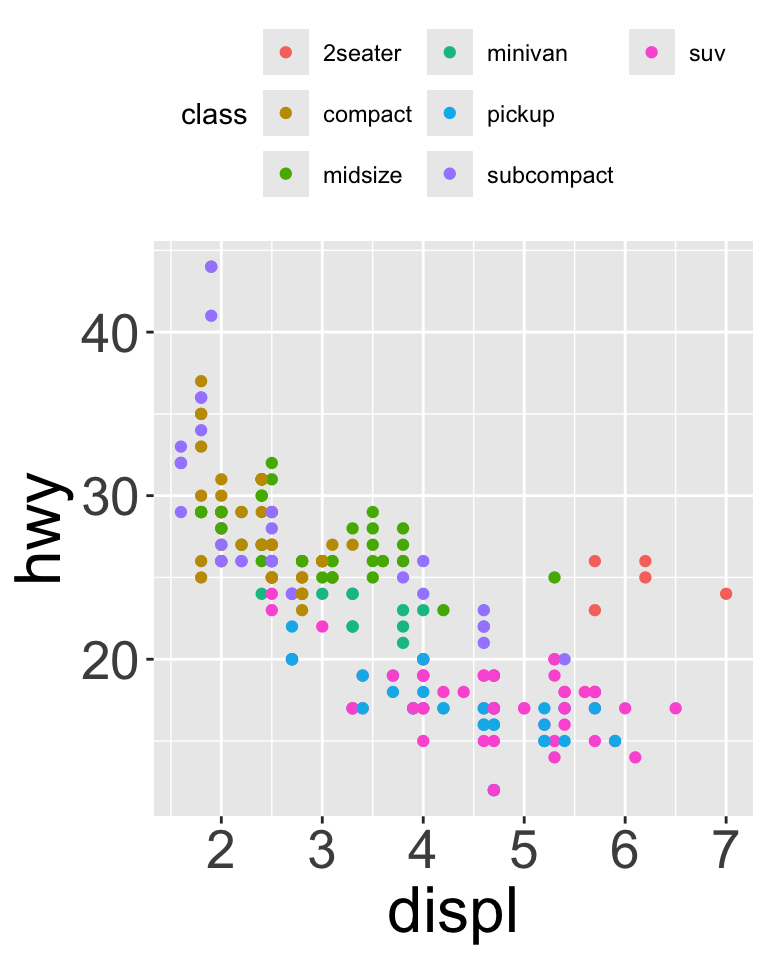
Legends
To control location of legend, use theme() (controls non data parts of a plot)
base <- ggplot(mpg, aes(x = displ, y = hwy)) + geom_point(aes(color = class))
base + theme(legend.position = "right") # the default
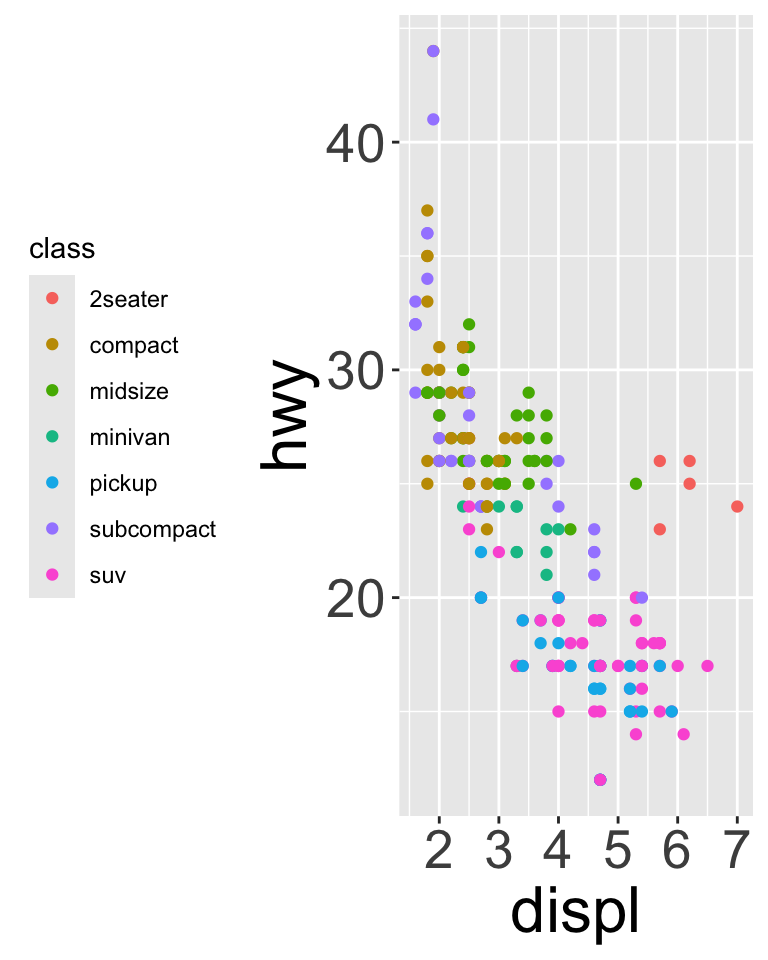
base + theme(legend.position = "left")
base + theme(legend.position = "top") + guides(color = guide_legend(nrow = 3))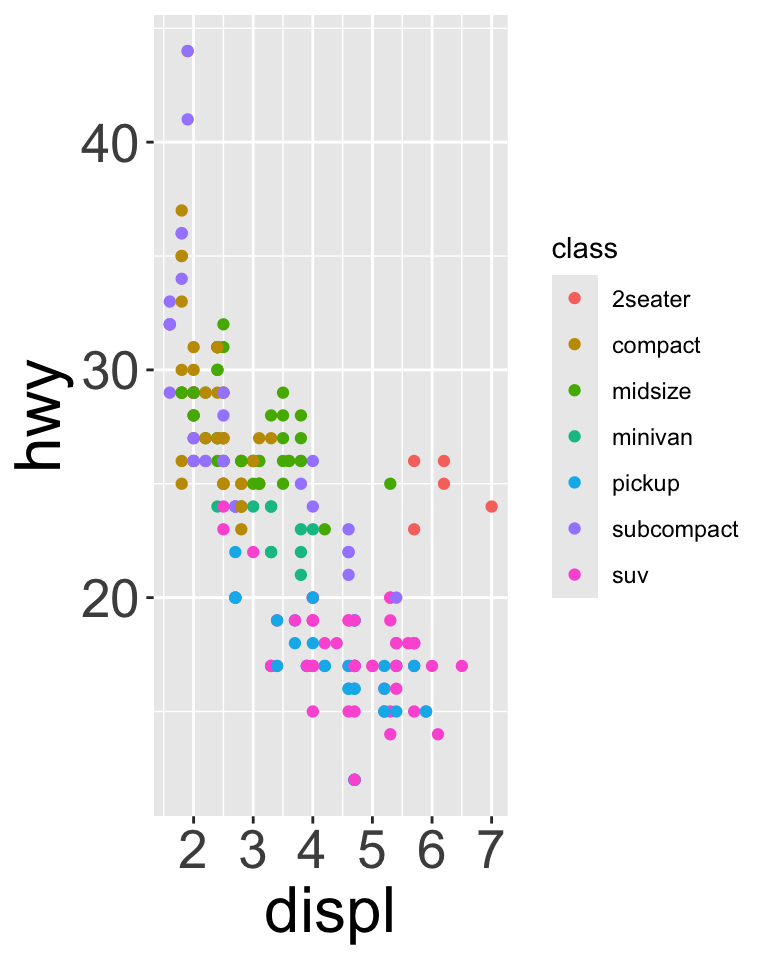
Replacing scales
Replacing scales
- Can use
scale_x_log10()andscale_y_log190)to plot regular values, but with ticks that are spread using a log scale
Replacing scales
- Color scales are also often replaced; especially using
scale_color_brewer()
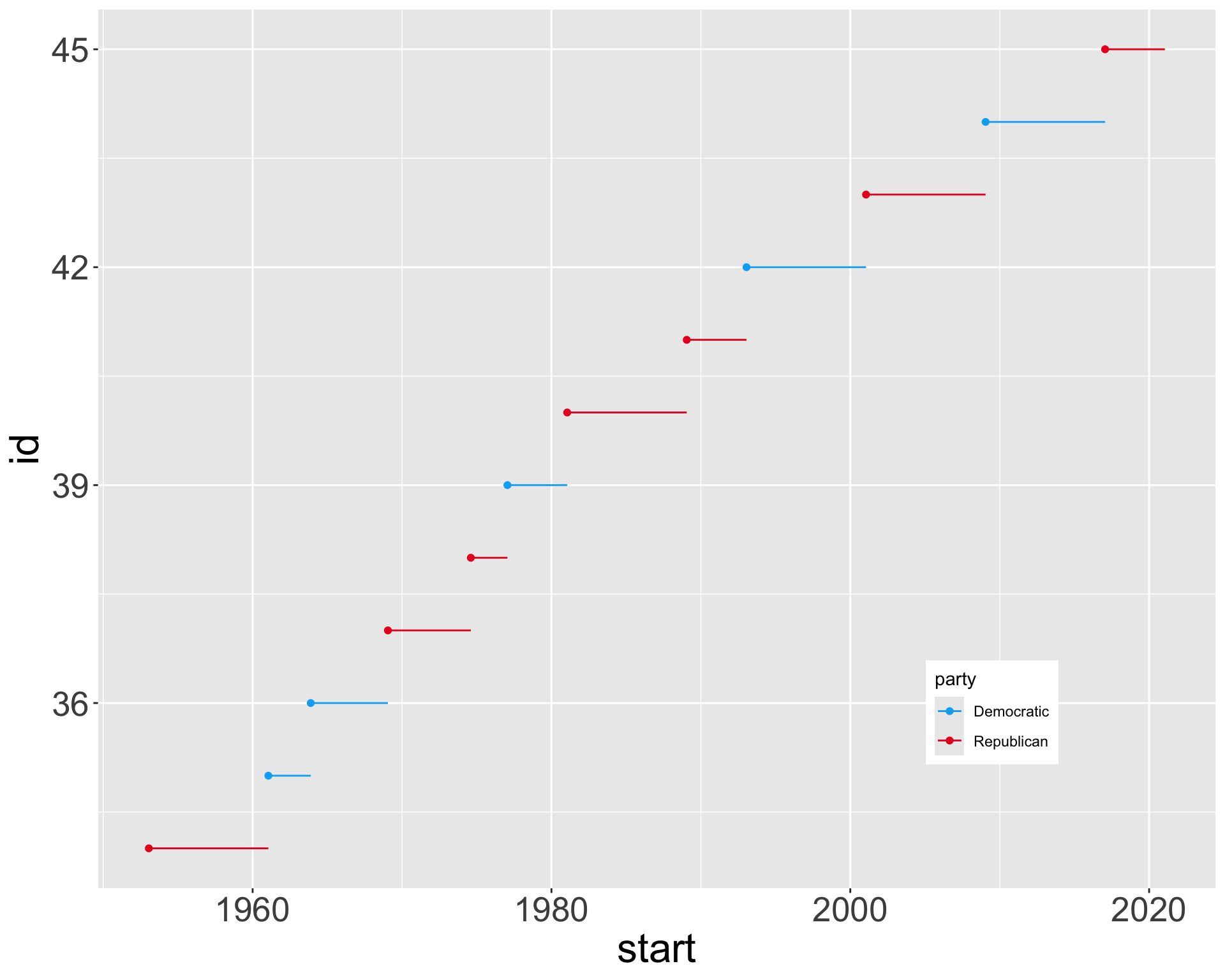
Replacing scales
Manual color scaling
scale_color_manual() allows giving specific colors for groups in the tibble
# A tibble: 6 × 4
name start end party
<chr> <date> <date> <chr>
1 Eisenhower 1953-01-20 1961-01-20 Republican
2 Kennedy 1961-01-20 1963-11-22 Democratic
3 Johnson 1963-11-22 1969-01-20 Democratic
4 Nixon 1969-01-20 1974-08-09 Republican
5 Ford 1974-08-09 1977-01-20 Republican
6 Carter 1977-01-20 1981-01-20 Democratic- Takes argument
values: a named vector, with entries of the formgroup=color. colorcan be either English names (“blue”, “red”) or hexadecimal color codes